The Challenge of Stability in High-Throughput Gene Expression Analysis: Comprehensive Selection and Evaluation of Reference Genes for BALB/c Mice Spleen Samples in the Leishmania infantum Infection Model
- PMID: 27668434
- PMCID: PMC5036817
- DOI: 10.1371/journal.pone.0163219
The Challenge of Stability in High-Throughput Gene Expression Analysis: Comprehensive Selection and Evaluation of Reference Genes for BALB/c Mice Spleen Samples in the Leishmania infantum Infection Model
Abstract
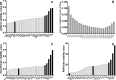
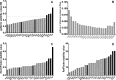
The interaction of Leishmania with BALB/c mice induces dramatic changes in transcriptome patterns in the parasite, but also in the target organs (spleen, liver…) due to its response against infection. Real-time quantitative PCR (qPCR) is an interesting approach to analyze these changes and understand the immunological pathways that lead to protection or progression of disease. However, qPCR results need to be normalized against one or more reference genes (RG) to correct for non-specific experimental variation. The development of technical platforms for high-throughput qPCR analysis, and powerful software for analysis of qPCR data, have acknowledged the problem that some reference genes widely used due to their known or suspected "housekeeping" roles, should be avoided due to high expression variability across different tissues or experimental conditions. In this paper we evaluated the stability of 112 genes using three different algorithms: geNorm, NormFinder and RefFinder in spleen samples from BALB/c mice under different experimental conditions (control and Leishmania infantum-infected mice). Despite minor discrepancies in the stability ranking shown by the three methods, most genes show very similar performance as RG (either good or poor) across this massive data set. Our results show that some of the genes traditionally used as RG in this model (i.e. B2m, Polr2a and Tbp) are clearly outperformed by others. In particular, the combination of Il2rg + Itgb2 was identified among the best scoring candidate RG for every group of mice and every algorithm used in this experimental model. Finally, we have demonstrated that using "traditional" vs rationally-selected RG for normalization of gene expression data may lead to loss of statistical significance of gene expression changes when using large-scale platforms, and therefore misinterpretation of results. Taken together, our results highlight the need for a comprehensive, high-throughput search for the most stable reference genes in each particular experimental model.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

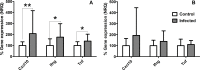
Similar articles
-
LEMming: A Linear Error Model to Normalize Parallel Quantitative Real-Time PCR (qPCR) Data as an Alternative to Reference Gene Based Methods.PLoS One. 2015 Sep 1;10(9):e0135852. doi: 10.1371/journal.pone.0135852. eCollection 2015. PLoS One. 2015. PMID: 26325269 Free PMC article.
-
Investigating reference genes for quantitative real-time PCR analysis across four chicken tissues.Gene. 2015 Apr 25;561(1):82-7. doi: 10.1016/j.gene.2015.02.016. Epub 2015 Feb 11. Gene. 2015. PMID: 25680290
-
Selection of reference genes for quantitative studies in acute myeloid leukaemia.Malays J Pathol. 2019 Dec;41(3):313-326. Malays J Pathol. 2019. PMID: 31901916
-
Selection of suitable reference genes for normalization of quantitative RT-PCR (RT-qPCR) expression data across twelve tissues of riverine buffaloes (Bubalus bubalis).PLoS One. 2018 Mar 6;13(3):e0191558. doi: 10.1371/journal.pone.0191558. eCollection 2018. PLoS One. 2018. PMID: 29509770 Free PMC article.
-
Identification of suitable reference genes in the mouse placenta.Placenta. 2016 Mar;39:7-15. doi: 10.1016/j.placenta.2015.12.017. Epub 2015 Dec 25. Placenta. 2016. PMID: 26992668
Cited by
-
Transcriptional Profiling of Immune-Related Genes in Leishmania infantum-Infected Mice: Identification of Potential Biomarkers of Infection and Progression of Disease.Front Cell Infect Microbiol. 2018 Jun 26;8:197. doi: 10.3389/fcimb.2018.00197. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30013952 Free PMC article.
-
Effect of 8-week of dietary micronutrient supplementation on gene expression in elite handball athletes.PLoS One. 2020 May 1;15(5):e0232237. doi: 10.1371/journal.pone.0232237. eCollection 2020. PLoS One. 2020. PMID: 32357196 Free PMC article. Clinical Trial.
-
The expression of immune response genes in patients with chronic Chagas disease is shifted toward the levels observed in healthy subjects as a result of treatment with Benznidazole.Front Cell Infect Microbiol. 2024 Jul 23;14:1439714. doi: 10.3389/fcimb.2024.1439714. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39119291 Free PMC article.
-
A systematic review on the selection of reference genes for gene expression studies in rodents: are the classics the best choice?Mol Biol Rep. 2024 Sep 26;51(1):1017. doi: 10.1007/s11033-024-09950-3. Mol Biol Rep. 2024. PMID: 39327364
-
Differential expression profile of genes involved in the immune response associated to progression of chronic Chagas disease.PLoS Negl Trop Dis. 2023 Jul 13;17(7):e0011474. doi: 10.1371/journal.pntd.0011474. eCollection 2023 Jul. PLoS Negl Trop Dis. 2023. PMID: 37440604 Free PMC article.
References
-
- World Health Organization. Control of the leishmaniases. World Health Organ Tech Rep Ser. 2010;949: xii–xiii, 1–186, back cover. Available: http://www.ncbi.nlm.nih.gov/pubmed/21485694 - PubMed
-
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6: 986–94. Available: http://www.ncbi.nlm.nih.gov/pubmed/8908518 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

