TSHZ3 deletion causes an autism syndrome and defects in cortical projection neurons
- PMID: 27668656
- PMCID: PMC5083212
- DOI: 10.1038/ng.3681
TSHZ3 deletion causes an autism syndrome and defects in cortical projection neurons
Abstract
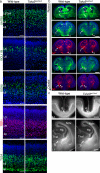
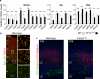
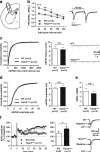
TSHZ3, which encodes a zinc-finger transcription factor, was recently positioned as a hub gene in a module of the genes with the highest expression in the developing human neocortex, but its functions remained unknown. Here we identify TSHZ3 as the critical region for a syndrome associated with heterozygous deletions at 19q12-q13.11, which includes autism spectrum disorder (ASD). In Tshz3-null mice, differentially expressed genes include layer-specific markers of cerebral cortical projection neurons (CPNs), and the human orthologs of these genes are strongly associated with ASD. Furthermore, mice heterozygous for Tshz3 show functional changes at synapses established by CPNs and exhibit core ASD-like behavioral abnormalities. These findings highlight essential roles for Tshz3 in CPN development and function, whose alterations can account for ASD in the newly defined TSHZ3 deletion syndrome.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
References for main text
-
- Association, A.P. Diagnostic and Statistical Manual of Mental Disorders, DSM-5. Fifth Edition American Psychiatric Association; Washington, DC: 2013.
Methods-only references
-
- Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. Second edition Academic Press; 2001.
-
- Banerjee-Basu S, Packer A. SFARI Gene: an evolving database for the autism research community. Dis. Models Mech. 2010;3:133–5. - PubMed
-
- Chassain C, et al. Metabolic, synaptic and behavioral impact of 5-week chronic deep brain stimulation in hemiparkinsonian rats. J. Neurochem. 2016;136:1004–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

