RNAi Screening of the Glucose-Regulated Chaperones in Cancer with Self-Assembled siRNA Nanostructures
- PMID: 27669096
- PMCID: PMC5378679
- DOI: 10.1021/acs.nanolett.6b02274
RNAi Screening of the Glucose-Regulated Chaperones in Cancer with Self-Assembled siRNA Nanostructures
Abstract
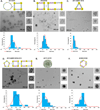
The emerging field of RNA nanotechnology has been used to design well-programmed, self-assembled nanostructures for applications in chemistry, biology, and medicine. At the forefront of its utility in cancer is the unrestricted ability to self-assemble multiple siRNAs within a single nanostructure formulation for the RNAi screening of a wide range of oncogenes while potentiating the gene therapy of malignant tumors. In our RNAi nanotechnology approach, V- and Y-shape RNA templates were designed and constructed for the self-assembly of discrete, higher-ordered siRNA nanostructures targeting the oncogenic glucose regulated chaperones. The GRP78-targeting siRNAs self-assembled into genetically encoded spheres, triangles, squares, pentagons and hexagons of discrete sizes and shapes according to TEM imaging. Furthermore, gel electrophoresis, thermal denaturation, and CD spectroscopy validated the prerequisite siRNA hybrids for their RNAi application. In a 24 sample siRNA screen conducted within the AN3CA endometrial cancer cells known to overexpress oncogenic GRP78 activity, the self-assembled siRNAs targeting multiple sites of GRP78 expression demonstrated more potent and long-lasting anticancer activity relative to their linear controls. Extending the scope of our RNAi screening approach, the self-assembled siRNA hybrids (5 nM) targeting of GRP-75, 78, and 94 resulted in significant (50-95%) knockdown of the glucose regulated chaperones, which led to synergistic effects in tumor cell cycle arrest (50-80%) and death (50-60%) within endometrial (AN3CA), cervical (HeLa), and breast (MDA-MB-231) cancer cell lines. Interestingly, a nontumorigenic lung (MRC5) cell line displaying normal glucose regulated chaperone levels was found to tolerate siRNA treatment and demonstrated less toxicity (5-20%) relative to the cancer cells that were found to be addicted to glucose regulated chaperones. These remarkable self-assembled siRNA nanostructures may thus encompass a new class of potent siRNAs that may be useful in screening important oncogene targets while improving siRNA therapeutic efficacy and specificity in cancer.
Keywords: GRP; RNAi nanotechnology; cancer gene therapy; cervical and breast cancer; chaperones; endometrial; glucose regulated proteins; siRNA nanostructures.
Figures
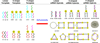



Similar articles
-
Enhanced Cancer Theranostics with Self-Assembled, Multilabeled siRNAs.ACS Omega. 2018 Oct 31;3(10):12975-12984. doi: 10.1021/acsomega.8b01999. Epub 2018 Oct 10. ACS Omega. 2018. PMID: 30411024 Free PMC article.
-
Direct Transfection of Fatty Acid Conjugated siRNAs and Knockdown of the Glucose-Regulated Chaperones in Prostate Cancer Cells.Bioconjug Chem. 2018 Nov 21;29(11):3638-3648. doi: 10.1021/acs.bioconjchem.8b00580. Epub 2018 Oct 10. Bioconjug Chem. 2018. PMID: 30235926
-
Antitumor therapeutic application of self-assembled RNAi-AuNP nanoconstructs: Combination of VEGF-RNAi and photothermal ablation.Theranostics. 2017 Jan 1;7(1):9-22. doi: 10.7150/thno.16042. eCollection 2017. Theranostics. 2017. PMID: 28042312 Free PMC article.
-
Human Papillomavirus: Current and Future RNAi Therapeutic Strategies for Cervical Cancer.J Clin Med. 2015 May 21;4(5):1126-55. doi: 10.3390/jcm4051126. J Clin Med. 2015. PMID: 26239469 Free PMC article. Review.
-
Polyethylenimines for RNAi-mediated gene targeting in vivo and siRNA delivery to the lung.Eur J Pharm Biopharm. 2011 Apr;77(3):438-49. doi: 10.1016/j.ejpb.2010.11.007. Epub 2010 Nov 18. Eur J Pharm Biopharm. 2011. PMID: 21093588 Review.
Cited by
-
Synthesis of native-like crosslinked duplex RNA and study of its properties.Bioorg Med Chem. 2017 Apr 1;25(7):2191-2199. doi: 10.1016/j.bmc.2017.02.034. Epub 2017 Feb 21. Bioorg Med Chem. 2017. PMID: 28268052 Free PMC article.
-
HSP70s in Breast Cancer: Promoters of Tumorigenesis and Potential Targets/Tools for Therapy.Cells. 2021 Dec 7;10(12):3446. doi: 10.3390/cells10123446. Cells. 2021. PMID: 34943954 Free PMC article. Review.
-
Size Matters: Arginine-Derived Peptides Targeting the PSMA Receptor Can Efficiently Complex but Not Transfect siRNA.Mol Ther Nucleic Acids. 2019 Dec 6;18:863-870. doi: 10.1016/j.omtn.2019.10.013. Epub 2019 Oct 23. Mol Ther Nucleic Acids. 2019. PMID: 31739211 Free PMC article.
-
Enhanced Cancer Theranostics with Self-Assembled, Multilabeled siRNAs.ACS Omega. 2018 Oct 31;3(10):12975-12984. doi: 10.1021/acsomega.8b01999. Epub 2018 Oct 10. ACS Omega. 2018. PMID: 30411024 Free PMC article.
-
Effects of Prolonged GRP78 Haploinsufficiency on Organ Homeostasis, Behavior, Cancer and Chemotoxic Resistance in Aged Mice.Sci Rep. 2017 Feb 1;7:40919. doi: 10.1038/srep40919. Sci Rep. 2017. PMID: 28145503 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

