Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta
- PMID: 27669148
- PMCID: PMC5115868
- DOI: 10.7554/eLife.17219
Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta
Abstract
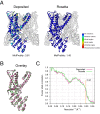
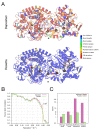
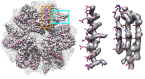
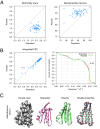
Cryo-EM has revealed the structures of many challenging yet exciting macromolecular assemblies at near-atomic resolution (3-4.5Å), providing biological phenomena with molecular descriptions. However, at these resolutions, accurately positioning individual atoms remains challenging and error-prone. Manually refining thousands of amino acids - typical in a macromolecular assembly - is tedious and time-consuming. We present an automated method that can improve the atomic details in models that are manually built in near-atomic-resolution cryo-EM maps. Applying the method to three systems recently solved by cryo-EM, we are able to improve model geometry while maintaining the fit-to-density. Backbone placement errors are automatically detected and corrected, and the refinement shows a large radius of convergence. The results demonstrate that the method is amenable to structures with symmetry, of very large size, and containing RNA as well as covalently bound ligands. The method should streamline the cryo-EM structure determination process, providing accurate and unbiased atomic structure interpretation of such maps.
Keywords: Rosetta; atomic models; biophysics; computational biology; cryo-EM; macromolecular assemblies; membrane proteins; none; structural biology; structure refinement; systems biology.
Conflict of interest statement
YS: Co-founder of Cyrus Biotechnology, Inc., which will develop and market graphic-interface software for using Rosetta. The other authors declare that no competing interests exist.
Figures




Similar articles
-
Iterative Molecular Dynamics-Rosetta Membrane Protein Structure Refinement Guided by Cryo-EM Densities.J Chem Theory Comput. 2017 Oct 10;13(10):5131-5145. doi: 10.1021/acs.jctc.7b00464. Epub 2017 Sep 26. J Chem Theory Comput. 2017. PMID: 28949136 Free PMC article.
-
Automated cryo-EM structure refinement using correlation-driven molecular dynamics.Elife. 2019 Mar 4;8:e43542. doi: 10.7554/eLife.43542. Elife. 2019. PMID: 30829573 Free PMC article.
-
Sample preparation of biological macromolecular assemblies for the determination of high-resolution structures by cryo-electron microscopy.Microscopy (Oxf). 2016 Feb;65(1):23-34. doi: 10.1093/jmicro/dfv367. Epub 2015 Dec 15. Microscopy (Oxf). 2016. PMID: 26671943 Review.
-
Accurate model annotation of a near-atomic resolution cryo-EM map.Proc Natl Acad Sci U S A. 2017 Mar 21;114(12):3103-3108. doi: 10.1073/pnas.1621152114. Epub 2017 Mar 7. Proc Natl Acad Sci U S A. 2017. PMID: 28270620 Free PMC article.
-
Current approaches for the fitting and refinement of atomic models into cryo-EM maps using CCP-EM.Acta Crystallogr D Struct Biol. 2018 Jun 1;74(Pt 6):492-505. doi: 10.1107/S2059798318007313. Epub 2018 May 30. Acta Crystallogr D Struct Biol. 2018. PMID: 29872001 Free PMC article. Review.
Cited by
-
Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.Nat Methods. 2020 Jul;17(7):699-707. doi: 10.1038/s41592-020-0878-9. Epub 2020 Jul 2. Nat Methods. 2020. PMID: 32616928 Free PMC article.
-
Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate.bioRxiv [Preprint]. 2020 Aug 6:2020.08.06.234674. doi: 10.1101/2020.08.06.234674. bioRxiv. 2020. Update in: Science. 2020 Nov 27;370(6520):1089-1094. doi: 10.1126/science.abe1502. PMID: 32793901 Free PMC article. Updated. Preprint.
-
Structural basis for the regulation of inositol trisphosphate receptors by Ca2+ and IP3.Nat Struct Mol Biol. 2018 Aug;25(8):660-668. doi: 10.1038/s41594-018-0089-6. Epub 2018 Jul 16. Nat Struct Mol Biol. 2018. PMID: 30013099 Free PMC article.
-
Cryo-EM structure of the activated RET signaling complex reveals the importance of its cysteine-rich domain.Sci Adv. 2019 Jul 31;5(7):eaau4202. doi: 10.1126/sciadv.aau4202. eCollection 2019 Jul. Sci Adv. 2019. PMID: 31392261 Free PMC article.
-
UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.Mol Cell. 2023 Aug 3;83(15):2753-2767.e10. doi: 10.1016/j.molcel.2023.06.028. Epub 2023 Jul 20. Mol Cell. 2023. PMID: 37478846 Free PMC article.
References
-
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica Section D Biological Crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

