Aerobic De-Epoxydation of Trichothecene Mycotoxins by a Soil Bacterial Consortium Isolated Using In Situ Soil Enrichment
- PMID: 27669304
- PMCID: PMC5086637
- DOI: 10.3390/toxins8100277
Aerobic De-Epoxydation of Trichothecene Mycotoxins by a Soil Bacterial Consortium Isolated Using In Situ Soil Enrichment
Abstract
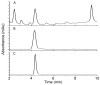
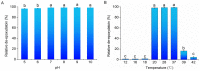
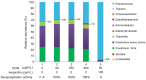
Globally, the trichothecene mycotoxins deoxynivalenol (DON) and nivalenol (NIV) are among the most widely distributed mycotoxins that contaminate small grain cereals. In this study, a bacterial consortium, PGC-3, with de-epoxydation activity was isolated from soil by an in situ soil enrichment method. Screening of 14 soil samples that were sprayed with DON revealed that 4 samples were able to biotransform DON into de-epoxydized DON (dE-DON). Among these, the PGC-3 consortium showed the highest and most stable activity to biotransform DON into dE-DON and NIV into dE-NIV. PGC-3 exhibited de-epoxydation activity at a wide range of pH (5-10) and temperatures (20-37 °C) values under aerobic conditions. Sequential subculturing with a continued exposure to DON substantially reduced the microbial population diversity of this consortium. Analyses of the 16S rDNA sequences indicated that PGC-3 comprised 10 bacterial genera. Among these, one species, Desulfitobacterium, showed a steady increase in relative abundance, from 0.03% to 1.55% (a 52-fold increase), as higher concentrations of DON were used in the subculture media, from 0 to 500 μg/mL. This study establishes the foundation to further develop bioactive agents that can detoxify trichothecene mycotoxins in cereals and enables for the characterization of detoxifying genes and their regulation.
Keywords: 16S rDNA sequencing; aerobic de-epoxydation; deoxynivalenol; soil bacterium; trichothecenes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Starkey D.E., Ward T.J., Aoki T., Gale L.R., Kistler H.C., Geiser D.M., Suga H., Toth B., Varga J., O’Donnell K. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 2007;44:1191–1204. doi: 10.1016/j.fgb.2007.03.001. - DOI - PubMed
-
- Ndoye M., Zhang J.B., Wang J.H., Gong A.D., Li H.P., Qu B., Li S.J., Liao Y.C. Nivalenol and 15-acetyldeoxynivalenol chemotypes of Fusarium graminearum clade species are prevalent on maize throughout China. J. Phytopathol. 2012;160:519–524. doi: 10.1111/j.1439-0434.2012.01944.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

