Membrane fission by dynamin: what we know and what we need to know
- PMID: 27670760
- PMCID: PMC5090216
- DOI: 10.15252/embj.201694613
Membrane fission by dynamin: what we know and what we need to know
Abstract
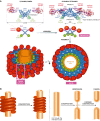
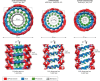
The large GTPase dynamin is the first protein shown to catalyze membrane fission. Dynamin and its related proteins are essential to many cell functions, from endocytosis to organelle division and fusion, and it plays a critical role in many physiological functions such as synaptic transmission and muscle contraction. Research of the past three decades has focused on understanding how dynamin works. In this review, we present the basis for an emerging consensus on how dynamin functions. Three properties of dynamin are strongly supported by experimental data: first, dynamin oligomerizes into a helical polymer; second, dynamin oligomer constricts in the presence of GTP; and third, dynamin catalyzes membrane fission upon GTP hydrolysis. We present the two current models for fission, essentially diverging in how GTP energy is spent. We further discuss how future research might solve the remaining open questions presently under discussion.
Keywords: GTPase; dynamin; endocytosis; membrane fission; molecular motor.
© 2016 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Figures


References
-
- Binns DD, Helms MK, Barylko B, Davis CT, Jameson DM, Albanesi JP, Eccleston JF (2000) The mechanism of GTP hydrolysis by dynamin II: a transient kinetic study. Biochemistry 39: 7188–7196 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

