Widespread natural variation of DNA methylation within angiosperms
- PMID: 27671052
- PMCID: PMC5037628
- DOI: 10.1186/s13059-016-1059-0
Widespread natural variation of DNA methylation within angiosperms
Abstract
Background: DNA methylation is an important feature of plant epigenomes, involved in the formation of heterochromatin and affecting gene expression. Extensive variation of DNA methylation patterns within a species has been uncovered from studies of natural variation. However, the extent to which DNA methylation varies between flowering plant species is still unclear. To understand the variation in genomic patterning of DNA methylation across flowering plant species, we compared single base resolution DNA methylomes of 34 diverse angiosperm species.
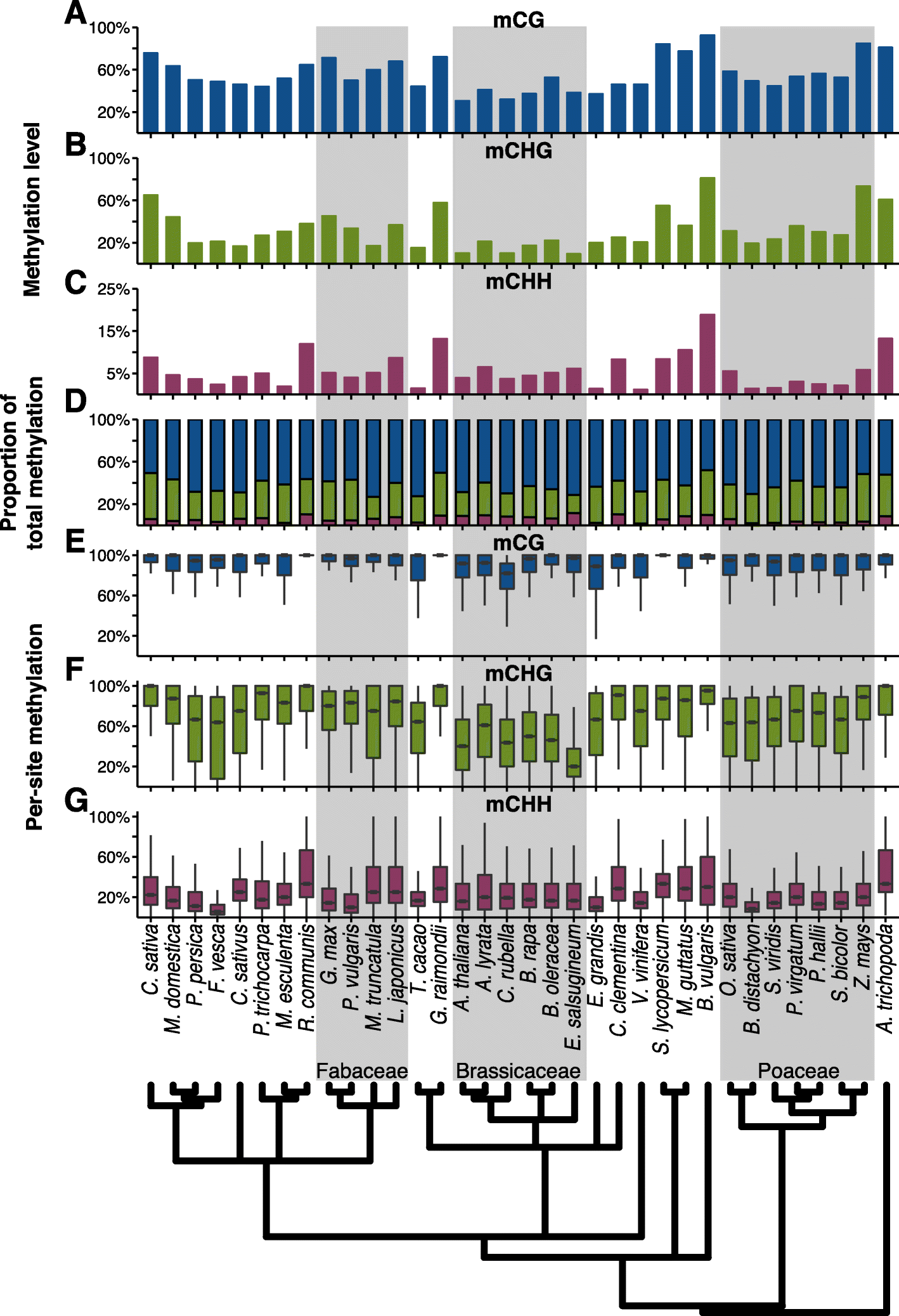
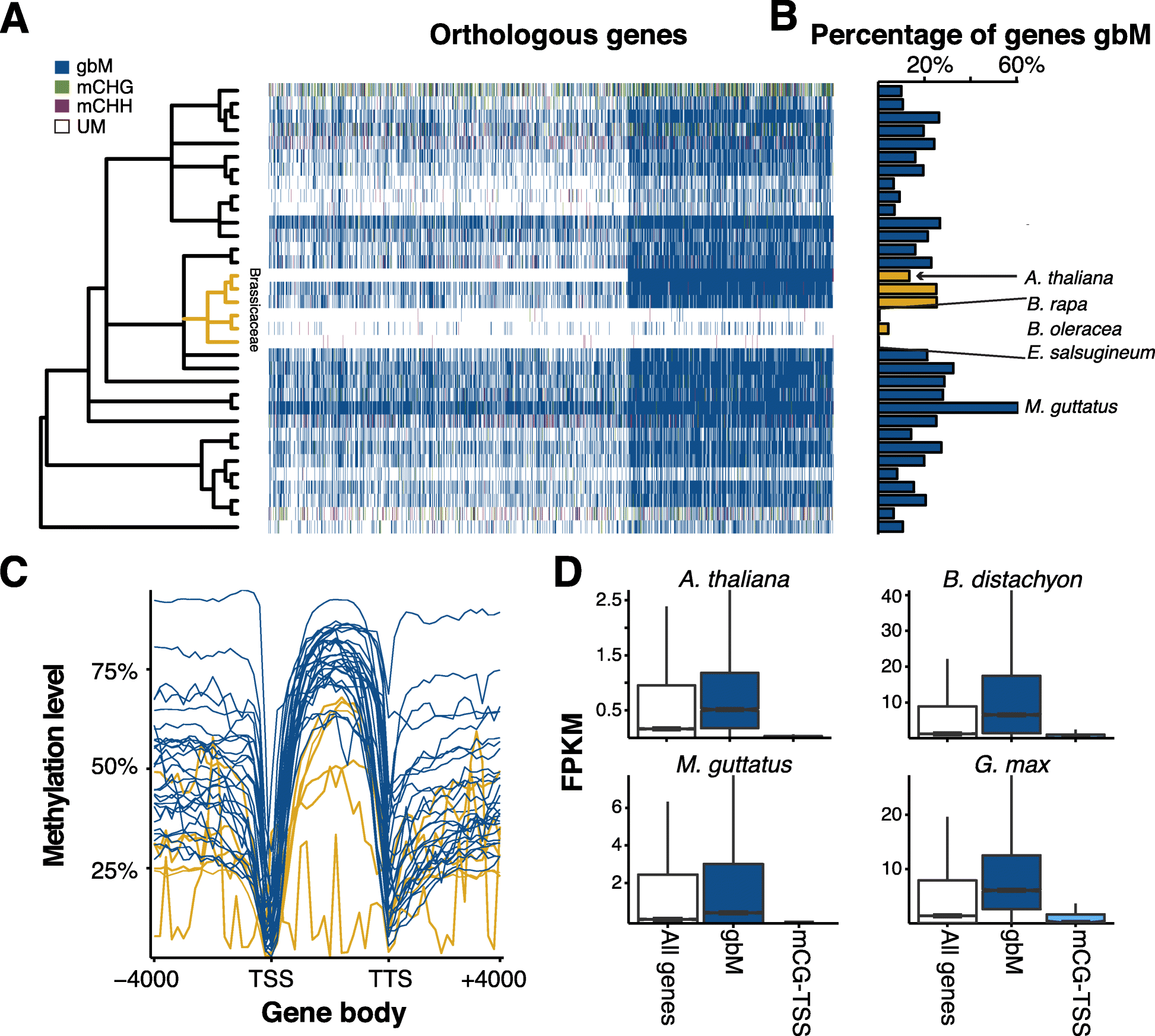
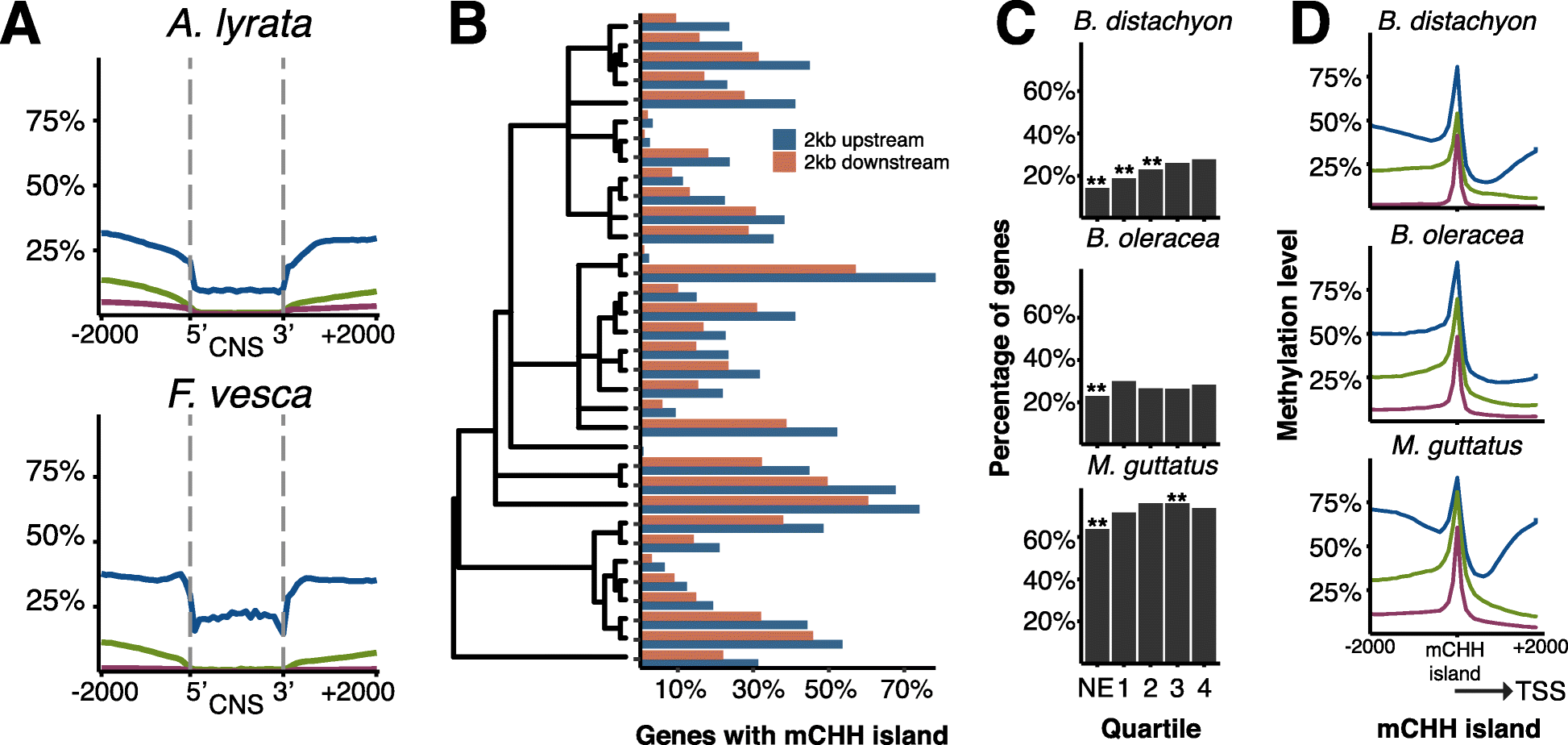
Results: By analyzing whole-genome bisulfite sequencing data in a phylogenetic context, it becomes clear that there is extensive variation throughout angiosperms in gene body DNA methylation, euchromatic silencing of transposons and repeats, as well as silencing of heterochromatic transposons. The Brassicaceae have reduced CHG methylation levels and also reduced or loss of CG gene body methylation. The Poaceae are characterized by a lack or reduction of heterochromatic CHH methylation and enrichment of CHH methylation in genic regions. Furthermore, low levels of CHH methylation are observed in a number of species, especially in clonally propagated species.
Conclusions: These results reveal the extent of variation in DNA methylation in angiosperms and show that DNA methylation patterns are broadly a reflection of the evolutionary and life histories of plant species.
Figures



Comment in
-
Prodigious plant methylomes.Genome Biol. 2016 Sep 27;17(1):197. doi: 10.1186/s13059-016-1065-2. Genome Biol. 2016. PMID: 27677311 Free PMC article.
References
-
- Colome-Tatche M, Cortijo S, Wardenaar R, Morgado L, Lahouze B, Sarazin A, et al. Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc Natl Acad Sci U S A. 2012;109(40):16240–5. doi: 10.1073/pnas.1212955109. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

