Global genetic diversity of Aedes aegypti
- PMID: 27671732
- PMCID: PMC5123671
- DOI: 10.1111/mec.13866
Global genetic diversity of Aedes aegypti
Abstract
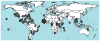
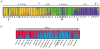
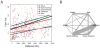
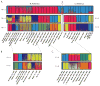
Mosquitoes, especially Aedes aegypti, are becoming important models for studying invasion biology. We characterized genetic variation at 12 microsatellite loci in 79 populations of Ae. aegypti from 30 countries in six continents, and used them to infer historical and modern patterns of invasion. Our results support the two subspecies Ae. aegypti formosus and Ae. aegypti aegypti as genetically distinct units. Ae. aegypti aegypti populations outside Africa are derived from ancestral African populations and are monophyletic. The two subspecies co-occur in both East Africa (Kenya) and West Africa (Senegal). In rural/forest settings (Rabai District of Kenya), the two subspecies remain genetically distinct, whereas in urban settings, they introgress freely. Populations outside Africa are highly genetically structured likely due to a combination of recent founder effects, discrete discontinuous habitats and low migration rates. Ancestral populations in sub-Saharan Africa are less genetically structured, as are the populations in Asia. Introduction of Ae. aegypti to the New World coinciding with trans-Atlantic shipping in the 16th to 18th centuries was followed by its introduction to Asia in the late 19th century from the New World or from now extinct populations in the Mediterranean Basin. Aedes mascarensis is a genetically distinct sister species to Ae. aegypti s.l. This study provides a reference database of genetic diversity that can be used to determine the likely origin of new introductions that occur regularly for this invasive species. The genetic uniqueness of many populations and regions has important implications for attempts to control Ae. aegypti, especially for the methods using genetic modification of populations.
Keywords: Aedes aegypti; Aedes mascarensis; history; invasion; microsatellites.
© 2016 John Wiley & Sons Ltd.
Figures



References
-
- Beebe NW, Ambrose L, Hill LA, Davis JB, Hapgood G, Cooper RD, Russell RC, Ritchie SA, reamer L, Lobo NF, Syafruddin D, den Hurk AF. Tracing the tiger: population genetics provides valuable insights into the Aedes (Stegomyia) albopictus invation of the Australasian region. PLoS Negl Trop Dis. 2013;7:e2361. - PMC - PubMed
-
- Beserra EB, de Castro FP, Jr, dos Santos JW, da Santos TS, Fernandes CR. Biology and thermal exigency of Aedes aegypti (L.) (Diptera: Culicidae) from four bioclimatic localities of Paraiba. Neotrop Entomol. 2006;35(6):853–860. - PubMed
-
- Black WC, Bennett KE, Gorrochotegui-Escalante N, Barillas-Mury C, Fernandez-Sala I, Munoz MDL, Farfan-Ale JA, Olson KE, Beaty BJ. Flavivirus susceptibility in Aedes aegypti. Arch Med Res. 2002;33:379–388. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

