DNA methylation profile of triple negative breast cancer-specific genes comparing lymph node positive patients to lymph node negative patients
- PMID: 27671774
- PMCID: PMC5037364
- DOI: 10.1038/srep33435
DNA methylation profile of triple negative breast cancer-specific genes comparing lymph node positive patients to lymph node negative patients
Abstract
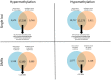
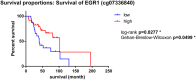
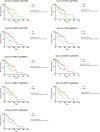
Triple negative breast cancer (TNBC) is the most aggressive breast cancer subtype with no targeted treatment available. Our previous study identified 38 TNBC-specific genes with altered expression comparing tumour to normal samples. This study aimed to establish whether DNA methylation contributed to these expression changes in the same cohort as well as disease progression from primary breast tumour to lymph node metastasis associated with changes in the epigenome. We obtained DNA from 23 primary TNBC samples, 12 matched lymph node metastases, and 11 matched normal adjacent tissues and assayed for differential methylation profiles using Illumina HumanMethylation450 BeadChips. The results were validated in an independent cohort of 70 primary TNBC samples. The expression of 16/38 TNBC-specific genes was associated with alteration in DNA methylation. Novel methylation changes between primary tumours and lymph node metastases, as well as those associated with survival were identified. Altered methylation of 18 genes associated with lymph node metastasis were identified and validated. This study reveals the important role DNA methylation plays in altered gene expression of TNBC-specific genes and lymph node metastases. The novel insights into progression of TNBC to secondary disease may provide potential prognostic indicators for this hard-to-treat breast cancer subtype.
Figures

References
-
- Carotenuto P. et al. Triple negative breast cancer: from molecular portrait to therapeutic intervention. Crit Rev Eukaryot Gene Expr 20, 17–34 (2010). - PubMed
-
- Wong-Brown M. W. et al. Prevalence of BRCA1 and BRCA2 germline mutations in patients with triple-negative breast cancer. Breast Cancer Res Treat 150, 71–80 (2015). - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

