Concerted effects of heterogeneous nuclear ribonucleoprotein C1/C2 to control vitamin D-directed gene transcription and RNA splicing in human bone cells
- PMID: 27672039
- PMCID: PMC5314791
- DOI: 10.1093/nar/gkw851
Concerted effects of heterogeneous nuclear ribonucleoprotein C1/C2 to control vitamin D-directed gene transcription and RNA splicing in human bone cells
Abstract
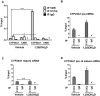
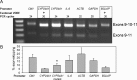
Traditionally recognized as an RNA splicing regulator, heterogeneous nuclear ribonucleoprotein C1/C2 (hnRNPC1/C2) can also bind to double-stranded DNA and function in trans as a vitamin D response element (VDRE)-binding protein. As such, hnRNPC1/C2 may couple transcription induced by the active form of vitamin D, 1,25-dihydroxyvitamin D (1,25(OH)2D) with subsequent RNA splicing. In MG63 osteoblastic cells, increased expression of the 1,25(OH)2D target gene CYP24A1 involved immunoprecipitation of hnRNPC1/C2 with CYP24A1 chromatin and RNA. Knockdown of hnRNPC1/C2 suppressed expression of CYP24A1, but also increased expression of an exon 10-skipped CYP24A1 splice variant; in a minigene model the latter was attenuated by a functional VDRE in the CYP24A1 promoter. In genome-wide analyses, knockdown of hnRNPC1/C2 resulted in 3500 differentially expressed genes and 2232 differentially spliced genes, with significant commonality between groups. 1,25(OH)2D induced 324 differentially expressed genes, with 187 also observed following hnRNPC1/C2 knockdown, and a further 168 unique to hnRNPC1/C2 knockdown. However, 1,25(OH)2D induced only 10 differentially spliced genes, with no overlap with differentially expressed genes. These data indicate that hnRNPC1/C2 binds to both DNA and RNA and influences both gene expression and RNA splicing, but these actions do not appear to be linked through 1,25(OH)2D-mediated induction of transcription.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Auboeuf D., Honig A., Berget S.M., O'Malley B.W. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. - PubMed
-
- Kwek K.Y., Murphy S., Furger A., Thomas B., O'Gorman W., Kimura H., Proudfoot N.J., Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 2002;9:800–805. - PubMed
-
- Tian H. RNA ligands generated against complex nuclear targets indicate a role for U1 snRNP in co-ordinating transcription and RNA splicing. FEBS Lett. 2001;509:282–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

