Trigger phosphodiesterases as a novel class of c-di-GMP effector proteins
- PMID: 27672149
- PMCID: PMC5052742
- DOI: 10.1098/rstb.2015.0498
Trigger phosphodiesterases as a novel class of c-di-GMP effector proteins
Abstract
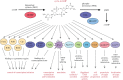
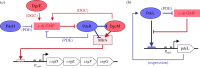
The bacterial second messenger c-di-GMP controls bacterial biofilm formation, motility, cell cycle progression, development and virulence. It is synthesized by diguanylate cyclases (with GGDEF domains), degraded by specific phosphodiesterases (PDEs, with EAL of HD-GYP domains) and sensed by a wide variety of c-di-GMP-binding effectors that control diverse targets. c-di-GMP-binding effectors can be riboswitches as well as proteins with highly diverse structures and functions. The latter include 'degenerate' GGDEF/EAL domain proteins that are enzymatically inactive but still able to bind c-di-GMP. Surprisingly, two enzymatically active 'trigger PDEs', the Escherichia coli proteins PdeR and PdeL, have recently been added to this list of c-di-GMP-sensing effectors. Mechanistically, trigger PDEs are multifunctional. They directly and specifically interact with a macromolecular target (e.g. with a transcription factor or directly with a promoter region), whose activity they control by their binding and degradation of c-di-GMP-their PDE activity thus represents the c-di-GMP sensor or effector function. In this process, c-di-GMP serves as a regulatory ligand, but in contrast to classical allosteric control, this ligand is also degraded. The resulting kinetics and circuitry of control are ideally suited for trigger PDEs to serve as key components in regulatory switches.This article is part of the themed issue 'The new bacteriology'.
Keywords: EAL domain; biofilm; cellulose; curli fibres; diguanylate cyclase; second messenger.
© 2016 The Authors.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

