Convergent evolution of pathogenicity islands in helper cos phage interference
- PMID: 27672154
- PMCID: PMC5052747
- DOI: 10.1098/rstb.2015.0505
Convergent evolution of pathogenicity islands in helper cos phage interference
Abstract
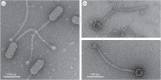
Staphylococcus aureus pathogenicity islands (SaPIs) are phage satellites that exploit the life cycle of their helper phages for their own benefit. Most SaPIs are packaged by their helper phages using a headful (pac) packaging mechanism. These SaPIs interfere with pac phage reproduction through a variety of strategies, including the redirection of phage capsid assembly to form small capsids, a process that depends on the expression of the SaPI-encoded cpmA and cpmB genes. Another SaPI subfamily is induced and packaged by cos-type phages, and although these cos SaPIs also block the life cycle of their inducing phages, the basis for this mechanism of interference remains to be deciphered. Here we have identified and characterized one mechanism by which the SaPIs interfere with cos phage reproduction. This mechanism depends on a SaPI-encoded gene, ccm, which encodes a protein involved in the production of small isometric capsids, compared with the prolate helper phage capsids. As the Ccm and CpmAB proteins are completely unrelated in sequence, this strategy represents a fascinating example of convergent evolution. Moreover, this result also indicates that the production of SaPI-sized particles is a widespread strategy of phage interference conserved during SaPI evolution.This article is part of the themed issue 'The new bacteriology'.
Keywords: PICIs; SaPIs; bacteriophage packaging; bacteriophage resistance; capsid morphogenesis; small capsids.
© 2016 The Authors.
Figures






References
-
- Ubeda C, Tormo MA, Cucarella C, Trotonda P, Foster TJ, Lasa I, Penadés JR. 2003. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol. Microbiol. 49, 193–210. (10.1046/j.1365-2958.2003.03577.x) - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
