Epigenomic profiling of primary gastric adenocarcinoma reveals super-enhancer heterogeneity
- PMID: 27677335
- PMCID: PMC5052795
- DOI: 10.1038/ncomms12983
Epigenomic profiling of primary gastric adenocarcinoma reveals super-enhancer heterogeneity
Abstract
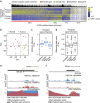
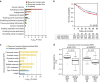
Regulatory enhancer elements in solid tumours remain poorly characterized. Here we apply micro-scale chromatin profiling to survey the distal enhancer landscape of primary gastric adenocarcinoma (GC), a leading cause of global cancer mortality. Integrating 110 epigenomic profiles from primary GCs, normal gastric tissues and cell lines, we highlight 36,973 predicted enhancers and 3,759 predicted super-enhancers respectively. Cell-line-defined super-enhancers can be subclassified by their somatic alteration status into somatic gain, loss and unaltered categories, each displaying distinct epigenetic, transcriptional and pathway enrichments. Somatic gain super-enhancers are associated with complex chromatin interaction profiles, expression patterns correlated with patient outcome and dense co-occupancy of the transcription factors CDX2 and HNF4α. Somatic super-enhancers are also enriched in genetic risk SNPs associated with cancer predisposition. Our results reveal a genome-wide reprogramming of the GC enhancer and super-enhancer landscape during tumorigenesis, contributing to dysregulated local and regional cancer gene expression.
Conflict of interest statement
W.F.O., S.L. and P.T are authors on patent applications entitled ‘Epigenomic Profiling of Primary Gastric Adenocarcinoma Reveals Super-Enhancer Heterogeneity', SG patent application no. 10201601141X (2016) and 10201606828P (2016). The remaining authors declare no competing financial interests.
Figures




References
-
- Zhang L. et al. Gene expression profiles in normal and cancer cells. Science 276, 1268–1272 (1997). - PubMed
-
- Spitz F. & Furlong E. E. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012). - PubMed
-
- Muratani M. et al. Nanoscale chromatin profiling of gastric adenocarcinoma reveals cancer-associated cryptic promoters and somatically acquired regulatory elements. Nat. Commun. 5, 4361 (2014). - PubMed
-
- Bernstein B. E. et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181 (2005). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

