Classifying lower grade glioma cases according to whole genome gene expression
- PMID: 27677590
- PMCID: PMC5342033
- DOI: 10.18632/oncotarget.12188
Classifying lower grade glioma cases according to whole genome gene expression
Abstract
Objective: To identify a gene-based signature as a novel prognostic model in lower grade gliomas.
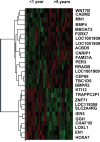
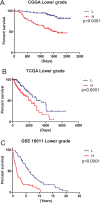
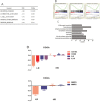
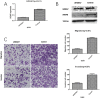
Results: A gene signature developed from HOXA7, SLC2A4RG and MN1 could segregate patients into low and high risk score groups with different overall survival (OS), and was validated in TCGA RNA-seq and GSE16011 mRNA array datasets. Receiver operating characteristic (ROC) was performed to show that the three-gene signature was more sensitive and specific than histology, grade, age, IDH1 mutation and 1p/19q co-deletion. Gene Set Enrichment Analysis (GSEA) and GO analysis showed high-risk samples were associated with tumor associated macrophages (TAMs) and highly invasive phenotypes. Moreover, HOXA7-siRNA inhibited migration and invasion in vitro, and downregulated MMP9 at the protein level in U251 glioma cells.
Methods: A cohort of 164 glioma specimens from the Chinese Glioma Genome Atlas (CGGA) array database were assessed as the training group. TCGA RNA-seq and GSE16011 mRNA array datasets were used for validation. Regression analyses and linear risk score assessment were performed for the identification of the three-gene signature comprising HOXA7, SLC2A4RG and MN1.
Co nclusions: We established a three-gene signature for lower grade gliomas, which could independently predict overall survival (OS) of lower grade glioma patients with higher sensitivity and specificity compared with other clinical characteristics. These findings indicate that the three-gene signature is a new prognostic model that could provide improved OS prediction and accurate therapies for lower grade glioma patients.
Keywords: gene signature; lower grade glioma; prognosis; risk score.
Conflict of interest statement
None for all authors.
Figures

References
-
- Wang Y, Wang X, Zhang J, Sun G, Luo H, Kang C, Pu P, Jiang T, Liu N, You Y. MicroRNAs involved in the EGFR/PTEN/AKT pathway in gliomas. Journal of neuro-oncology. 2012;106:217–224. - PubMed
-
- Yang P, You G, Zhang W, Wang Y, Wang Y, Yao K, Jiang T. Correlation of preoperative seizures with clinicopathological factors and prognosis in anaplastic gliomas: a report of 198 patients from China. Seizure. 2014;23:844–851. - PubMed
-
- Jiang T, Mao Y, Ma W, Mao Q, You Y, Yang X, Jiang C, Kang C, Li X, Chen L, Qiu X, Wang W, Li W, Yao Y, Li S, Li S, et al. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer letters. 2016;375:263–273. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

