Exposing the Three-Dimensional Biogeography and Metabolic States of Pathogens in Cystic Fibrosis Sputum via Hydrogel Embedding, Clearing, and rRNA Labeling
- PMID: 27677788
- PMCID: PMC5040109
- DOI: 10.1128/mBio.00796-16
Exposing the Three-Dimensional Biogeography and Metabolic States of Pathogens in Cystic Fibrosis Sputum via Hydrogel Embedding, Clearing, and rRNA Labeling
Abstract
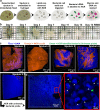
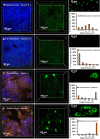
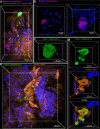
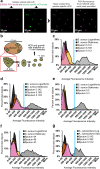
Physiological resistance to antibiotics confounds the treatment of many chronic bacterial infections, motivating researchers to identify novel therapeutic approaches. To do this effectively, an understanding of how microbes survive in vivo is needed. Though much can be inferred from bulk approaches to characterizing complex environments, essential information can be lost if spatial organization is not preserved. Here, we introduce a tissue-clearing technique, termed MiPACT, designed to retain and visualize bacteria with associated proteins and nucleic acids in situ on various spatial scales. By coupling MiPACT with hybridization chain reaction (HCR) to detect rRNA in sputum samples from cystic fibrosis (CF) patients, we demonstrate its ability to survey thousands of bacteria (or bacterial aggregates) over millimeter scales and quantify aggregation of individual species in polymicrobial communities. By analyzing aggregation patterns of four prominent CF pathogens, Staphylococcus aureus, Pseudomonas aeruginosa, Streptococcus sp., and Achromobacter xylosoxidans, we demonstrate a spectrum of aggregation states: from mostly single cells (A. xylosoxidans), to medium-sized clusters (S. aureus), to a mixture of single cells and large aggregates (P. aeruginosa and Streptococcus sp.). Furthermore, MiPACT-HCR revealed an intimate interaction between Streptococcus sp. and specific host cells. Lastly, by comparing standard rRNA fluorescence in situ hybridization signals to those from HCR, we found that different populations of S. aureus and A. xylosoxidans grow slowly overall yet exhibit growth rate heterogeneity over hundreds of microns. These results demonstrate the utility of MiPACT-HCR to directly capture the spatial organization and metabolic activity of bacteria in complex systems, such as human sputum.
Importance: The advent of metagenomic and metatranscriptomic analyses has improved our understanding of microbial communities by empowering us to identify bacteria, calculate their abundance, and profile gene expression patterns in complex environments. We are still technologically limited, however, in regards to the many questions that bulk measurements cannot answer, specifically in assessing the spatial organization of microbe-microbe and microbe-host interactions. Here, we demonstrate the power of an enhanced optical clearing method, MiPACT, to survey important aspects of bacterial physiology (aggregation, host interactions, and growth rate), in situ, with preserved spatial information when coupled to rRNA detection by HCR. Our application of MiPACT-HCR to cystic fibrosis patient sputum revealed species-specific aggregation patterns, yet slow growth characterized the vast majority of bacterial cells regardless of their cell type. More broadly, MiPACT, coupled with fluorescent labeling, promises to advance the direct study of microbial communities in diverse environments, including microbial habitats within mammalian systems.
Copyright © 2016 DePas et al.
Figures
References
-
- Jorth P, Staudinger BJ, Wu X, Hisert KB, Hayden H, Garudathri J, Harding CL, Radey MC, Rezayat A, Bautista G, Berrington WR, Goddard AF, Zheng C, Angermeyer A, Brittnacher MJ, Kitzman J, Shendure J, Fligner CL, Mittler J, Aitken ML, Manoil C, Bruce JE, Yahr TL, Singh PK. 2015. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe 18:307–319. doi:10.1016/j.chom.2015.07.006. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
