Assessing the Potential Effects of Active Site Mg2+ Ions in the glmS Ribozyme-Cofactor Complex
- PMID: 27677922
- PMCID: PMC5117136
- DOI: 10.1021/acs.jpclett.6b01854
Assessing the Potential Effects of Active Site Mg2+ Ions in the glmS Ribozyme-Cofactor Complex
Abstract
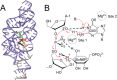
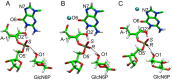
Ribozymes employ diverse catalytic strategies in their self-cleavage mechanisms, including the use of divalent metal ions. This work explores the effects of Mg2+ ions in the active site of the glmS ribozyme-GlcN6P cofactor complex using computational methods. Deleterious and potentially beneficial effects of an active site Mg2+ ion on the self-cleavage reaction were identified. The presence of a Mg2+ ion near the scissile phosphate oxygen atoms at the cleavage site was determined to be deleterious, and thereby anticatalytic, due to electrostatic repulsion of the cofactor, disruption of key hydrogen-bonding interactions, and obstruction of nucleophilic attack. On the other hand, the presence of a Mg2+ ion at another position in the active site, the Hoogsteen face of the putative base, was found to avoid these deleterious effects and to be potentially catalytically favorable owing to the stabilization of negative charge and pKa shifting of the guanine base.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Activation of the glmS Ribozyme Nucleophile via Overdetermined Hydrogen Bonding.Biochemistry. 2017 Aug 22;56(33):4313-4317. doi: 10.1021/acs.biochem.7b00662. Epub 2017 Aug 8. Biochemistry. 2017. PMID: 28787138
-
Chemical feasibility of the general acid/base mechanism of glmS ribozyme self-cleavage.Biopolymers. 2015 Oct;103(10):550-62. doi: 10.1002/bip.22657. Biopolymers. 2015. PMID: 25858644 Free PMC article.
-
A divalent cation-dependent variant of the glmS ribozyme with stringent Ca2+ selectivity co-opts a preexisting nonspecific metal ion-binding site.RNA. 2017 Mar;23(3):355-364. doi: 10.1261/rna.059824.116. Epub 2016 Dec 8. RNA. 2017. PMID: 27932587 Free PMC article.
-
Catalytic strategies of self-cleaving ribozymes.Acc Chem Res. 2008 Aug;41(8):1027-35. doi: 10.1021/ar800050c. Epub 2008 Jul 25. Acc Chem Res. 2008. PMID: 18652494 Review.
-
Structural and Biochemical Properties of Novel Self-Cleaving Ribozymes.Molecules. 2017 Apr 24;22(4):678. doi: 10.3390/molecules22040678. Molecules. 2017. PMID: 28441772 Free PMC article. Review.
Cited by
-
Graph deep learning locates magnesium ions in RNA.QRB Discov. 2022;3:e20. doi: 10.1017/qrd.2022.17. Epub 2022 Oct 6. QRB Discov. 2022. PMID: 37292390 Free PMC article.
-
Chemically Accurate Relative Folding Stability of RNA Hairpins from Molecular Simulations.J Chem Theory Comput. 2018 Dec 11;14(12):6598-6612. doi: 10.1021/acs.jctc.8b00633. Epub 2018 Nov 27. J Chem Theory Comput. 2018. PMID: 30375860 Free PMC article.
-
Simulations predict preferred Mg2+ coordination in a nonenzymatic primer-extension reaction center.Biophys J. 2024 Jun 18;123(12):1579-1591. doi: 10.1016/j.bpj.2024.04.032. Epub 2024 May 3. Biophys J. 2024. PMID: 38702884 Free PMC article.
-
Sodium and Magnesium Ion Location at the Backbone and at the Nucleobase of RNA: Ab Initio Molecular Dynamics in Water Solution.ACS Omega. 2022 Jun 23;7(27):23234-23244. doi: 10.1021/acsomega.2c01327. eCollection 2022 Jul 12. ACS Omega. 2022. PMID: 35847262 Free PMC article.
-
RNA Structural Dynamics As Captured by Molecular Simulations: A Comprehensive Overview.Chem Rev. 2018 Apr 25;118(8):4177-4338. doi: 10.1021/acs.chemrev.7b00427. Epub 2018 Jan 3. Chem Rev. 2018. PMID: 29297679 Free PMC article. Review.
References
-
- Wedekind J. E.Metal Ion Binding and Function in Natural and Artificial Small RNA Enzymes from a Structural Perspective. Structural and Catalytic Roles of Metal Ions in RNA; Metal Ions Life Sciences; Royal Society of Chemistry: Cambridge, U.K., 2011; Vol. 9; pp 299–345. DOI: 10.1039/9781849732512-00299 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

