Structural and Functional Characterization of a Ruminal β-Glycosidase Defines a Novel Subfamily of Glycoside Hydrolase Family 3 with Permuted Domain Topology
- PMID: 27679487
- PMCID: PMC5104943
- DOI: 10.1074/jbc.M116.747527
Structural and Functional Characterization of a Ruminal β-Glycosidase Defines a Novel Subfamily of Glycoside Hydrolase Family 3 with Permuted Domain Topology
Abstract
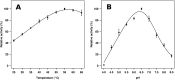
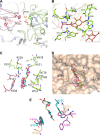
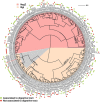
Metagenomics has opened up a vast pool of genes for putative, yet uncharacterized, enzymes. It widens our knowledge on the enzyme diversity world and discloses new families for which a clear classification is still needed, as is exemplified by glycoside hydrolase family-3 (GH3) proteins. Herein, we describe a GH3 enzyme (GlyA1) from resident microbial communities in strained ruminal fluid. The enzyme is a β-glucosidase/β-xylosidase that also shows β-galactosidase, β-fucosidase, α-arabinofuranosidase, and α-arabinopyranosidase activities. Short cello- and xylo-oligosaccharides, sophorose and gentibiose, are among the preferred substrates, with the large polysaccharide lichenan also being hydrolyzed by GlyA1 The determination of the crystal structure of the enzyme in combination with deletion and site-directed mutagenesis allowed identification of its unusual domain composition and the active site architecture. Complexes of GlyA1 with glucose, galactose, and xylose allowed picturing the catalytic pocket and illustrated the molecular basis of the substrate specificity. A hydrophobic platform defined by residues Trp-711 and Trp-106, located in a highly mobile loop, appears able to allocate differently β-linked bioses. GlyA1 includes an additional C-terminal domain previously unobserved in GH3 members, but crystallization of the full-length enzyme was unsuccessful. Therefore, small angle x-ray experiments have been performed to investigate the molecular flexibility and overall putative shape. This study provided evidence that GlyA1 defines a new subfamily of GH3 proteins with a novel permuted domain topology. Phylogenetic analysis indicates that this topology is associated with microbes inhabiting the digestive tracts of ruminants and other animals, feeding on chemically diverse plant polymeric materials.
Keywords: GH3 family; X-ray crystallography; enzyme structure; enzymology; glycoside hydrolase; metagenomics; permuted domains topology; phylogenetic analysis; protein structure; β-glycosidase.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

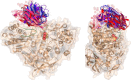
Similar articles
-
The structure of a glycoside hydrolase 29 family member from a rumen bacterium reveals unique, dual carbohydrate-binding domains.Acta Crystallogr F Struct Biol Commun. 2016 Oct 1;72(Pt 10):750-761. doi: 10.1107/S2053230X16014072. Epub 2016 Sep 22. Acta Crystallogr F Struct Biol Commun. 2016. PMID: 27710940 Free PMC article.
-
Structural and functional analyses of beta-glucosidase 3B from Thermotoga neapolitana: a thermostable three-domain representative of glycoside hydrolase 3.J Mol Biol. 2010 Apr 2;397(3):724-39. doi: 10.1016/j.jmb.2010.01.072. Epub 2010 Feb 6. J Mol Biol. 2010. PMID: 20138890
-
Discovery of α-l-arabinopyranosidases from human gut microbiome expands the diversity within glycoside hydrolase family 42.J Biol Chem. 2017 Dec 22;292(51):21092-21101. doi: 10.1074/jbc.M117.792598. Epub 2017 Oct 23. J Biol Chem. 2017. PMID: 29061847 Free PMC article.
-
Rational protein design for thermostabilization of glycoside hydrolases based on structural analysis.Appl Microbiol Biotechnol. 2018 Oct;102(20):8677-8684. doi: 10.1007/s00253-018-9288-7. Epub 2018 Aug 14. Appl Microbiol Biotechnol. 2018. PMID: 30109396 Review.
-
Peculiarities and systematics of microbial diglycosidases, and their applications in food technology.Appl Microbiol Biotechnol. 2021 Apr;105(7):2693-2700. doi: 10.1007/s00253-021-11219-9. Epub 2021 Mar 21. Appl Microbiol Biotechnol. 2021. PMID: 33745010 Review.
Cited by
-
Structural and mutational analysis of glycoside hydrolase family 1 Br2 β-glucosidase derived from bovine rumen metagenome.Heliyon. 2023 Nov 7;9(11):e21923. doi: 10.1016/j.heliyon.2023.e21923. eCollection 2023 Nov. Heliyon. 2023. PMID: 38034805 Free PMC article.
-
Crystal structure and identification of amino acid residues for catalysis and binding of GH3 AnBX β-xylosidase from Aspergillus niger.Appl Microbiol Biotechnol. 2023 Apr;107(7-8):2335-2349. doi: 10.1007/s00253-023-12445-z. Epub 2023 Mar 6. Appl Microbiol Biotechnol. 2023. PMID: 36877249
-
Functional metagenomics reveals novel β-galactosidases not predictable from gene sequences.PLoS One. 2017 Mar 8;12(3):e0172545. doi: 10.1371/journal.pone.0172545. eCollection 2017. PLoS One. 2017. PMID: 28273103 Free PMC article.
-
Identification and Immobilization of an Invertase With High Specific Activity and Sucrose Tolerance Ability of Gongronella sp. w5 for High Fructose Syrup Preparation.Front Microbiol. 2020 Apr 9;11:633. doi: 10.3389/fmicb.2020.00633. eCollection 2020. Front Microbiol. 2020. PMID: 32328053 Free PMC article.
-
Structure and evolutionary trace-assisted screening of a residue swapping the substrate ambiguity and chiral specificity in an esterase.Comput Struct Biotechnol J. 2021 Apr 18;19:2307-2317. doi: 10.1016/j.csbj.2021.04.041. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33995922 Free PMC article.
References
-
- Lee J. H., Hyun Y. J., and Kim D. H. (2011) Cloning and characterization of α-l-arabinofuranosidase and bifunctional α-l-arabinopyranosidase/β-d-galactopyranosidase from Bifidobacterium longum H-1. J. Appl. Microbiol. 111, 1097–1107 - PubMed
-
- Mayer C., Vocadlo D. J., Mah M., Rupitz K., Stoll D., Warren R. A., and Withers S. G. (2006) Characterization of a β-N-acetylhexosaminidase and a β-N-acetylglucosaminidase/β-glucosidase from Cellulomonas fimi. FEBS J. 273, 2929–2941 - PubMed
-
- DeBoy R. T., Mongodin E. F., Fouts D. E., Tailford L. E., Khouri H., Emerson J. B., Mohamoud Y., Watkins K., Henrissat B., Gilbert H. J., and Nelson K. E. (2008) Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J. Bacteriol. 190, 5455–5463 - PMC - PubMed
-
- Mai V., Wiegel J., and Lorenz W. W. (2000) Cloning, sequencing, and characterization of the bifunctional xylosidase-arabinosidase from the anaerobic thermophile Thermoanaerobacter ethanolicus. Gene 247, 137–143 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

