Distinct Bacterial Communities in Surficial Seafloor Sediments Following the 2010 Deepwater Horizon Blowout
- PMID: 27679609
- PMCID: PMC5020131
- DOI: 10.3389/fmicb.2016.01384
Distinct Bacterial Communities in Surficial Seafloor Sediments Following the 2010 Deepwater Horizon Blowout
Abstract
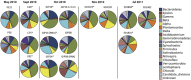
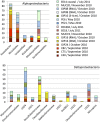
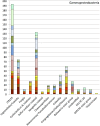
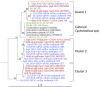
A major fraction of the petroleum hydrocarbons discharged during the 2010 Macondo oil spill became associated with and sank to the seafloor as marine snow flocs. This sedimentation pulse induced the development of distinct bacterial communities. Between May 2010 and July 2011, full-length 16S rRNA gene clone libraries demonstrated bacterial community succession in oil-polluted sediment samples near the wellhead area. Libraries from early May 2010, before the sedimentation event, served as the baseline control. Freshly deposited oil-derived marine snow was collected on the surface of sediment cores in September 2010, and was characterized by abundantly detected members of the marine Roseobacter cluster within the Alphaproteobacteria. Samples collected in mid-October 2010 closest to the wellhead contained members of the sulfate-reducing, anaerobic bacterial families Desulfobacteraceae and Desulfobulbaceae within the Deltaproteobacteria, suggesting that the oil-derived sedimentation pulse triggered bacterial oxygen consumption and created patchy anaerobic microniches that favored sulfate-reducing bacteria. Phylotypes of the polycyclic aromatic hydrocarbon-degrading genus Cycloclasticus, previously found both in surface oil slicks and the deep hydrocarbon plume, were also found in oil-derived marine snow flocs sedimenting on the seafloor in September 2010, and in surficial sediments collected in October and November 2010, but not in any of the control samples. Due to the relative recalcitrance and stability of polycyclic aromatic compounds, Cycloclasticus represents the most persistent microbial marker of seafloor hydrocarbon deposition that we could identify in this dataset. The bacterial imprint of the DWH oil spill had diminished in late November 2010, when the bacterial communities in oil-impacted sediment samples collected near the Macondo wellhead began to resemble their pre-spill counterparts and spatial controls. Samples collected in summer of 2011 did not show a consistent bacterial community signature, suggesting that the bacterial community was no longer shaped by the DWH fallout of oil-derived marine snow, but instead by location-specific and seasonal factors.
Keywords: Cycloclasticus; Deepwater Horizon; MOSSFA; bacterial populations; marine sediment; marine snow.
Figures



Similar articles
-
Enrichment of Fusobacteria in Sea Surface Oil Slicks from the Deepwater Horizon Oil Spill.Microorganisms. 2016 Jul 27;4(3):24. doi: 10.3390/microorganisms4030024. Microorganisms. 2016. PMID: 27681918 Free PMC article.
-
Hydrocarbons in Deep-Sea Sediments following the 2010 Deepwater Horizon Blowout in the Northeast Gulf of Mexico.PLoS One. 2015 May 28;10(5):e0128371. doi: 10.1371/journal.pone.0128371. eCollection 2015. PLoS One. 2015. PMID: 26020923 Free PMC article.
-
Hydrocarbon degradation and response of seafloor sediment bacterial community in the northern Gulf of Mexico to light Louisiana sweet crude oil.ISME J. 2018 Oct;12(10):2532-2543. doi: 10.1038/s41396-018-0190-1. Epub 2018 Jun 27. ISME J. 2018. PMID: 29950702 Free PMC article.
-
Was the extreme and wide-spread marine oil-snow sedimentation and flocculent accumulation (MOSSFA) event during the Deepwater Horizon blow-out unique?Mar Pollut Bull. 2015 Nov 15;100(1):5-12. doi: 10.1016/j.marpolbul.2015.08.023. Epub 2015 Sep 8. Mar Pollut Bull. 2015. PMID: 26359115 Review.
-
A critical review of marine snow in the context of oil spills and oil spill dispersant treatment with focus on the Deepwater Horizon oil spill.Mar Pollut Bull. 2018 Oct;135:346-356. doi: 10.1016/j.marpolbul.2018.07.028. Epub 2018 Jul 18. Mar Pollut Bull. 2018. PMID: 30301046 Review.
Cited by
-
In situ microcosms deployed at the coast of British Columbia (Canada) to study dilbit weathering and associated microbial communities under marine conditions.FEMS Microbiol Ecol. 2021 Jun 18;97(7):fiab082. doi: 10.1093/femsec/fiab082. FEMS Microbiol Ecol. 2021. PMID: 34124756 Free PMC article.
-
Influence of oil, dispersant, and pressure on microbial communities from the Gulf of Mexico.Sci Rep. 2020 Apr 27;10(1):7079. doi: 10.1038/s41598-020-63190-6. Sci Rep. 2020. PMID: 32341378 Free PMC article.
-
Enrichment of Fusobacteria in Sea Surface Oil Slicks from the Deepwater Horizon Oil Spill.Microorganisms. 2016 Jul 27;4(3):24. doi: 10.3390/microorganisms4030024. Microorganisms. 2016. PMID: 27681918 Free PMC article.
-
The Variable Influence of Dispersant on Degradation of Oil Hydrocarbons in Subarctic Deep-Sea Sediments at Low Temperatures (0-5 °C).Sci Rep. 2017 May 22;7(1):2253. doi: 10.1038/s41598-017-02475-9. Sci Rep. 2017. PMID: 28533547 Free PMC article.
-
Anaerobic degradation of hexadecane and phenanthrene coupled to sulfate reduction by enriched consortia from northern Gulf of Mexico seafloor sediment.Sci Rep. 2019 Feb 4;9(1):1239. doi: 10.1038/s41598-018-36567-x. Sci Rep. 2019. PMID: 30718896 Free PMC article.
References
-
- Arnosti C., Ziervogel K., Yang T., Teske A. (2016). Oil-derived marine aggregates - hot spots of polysaccharide degradation by specialized bacterial communities. Deep Sea Res. Part II 129 179–186. 10.1016/j.dsr2.2014.12.008 - DOI
-
- Ashelford K. E., Chuzhanova N. A., Fry J. C., Jones A. J., Weightman A. J. (2005). At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71 7724–7736. 10.1128/AEM.71.12.7724-7736.2005 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

