Genome-wide identification of multifunctional laccase gene family in cotton (Gossypium spp.); expression and biochemical analysis during fiber development
- PMID: 27679939
- PMCID: PMC5041144
- DOI: 10.1038/srep34309
Genome-wide identification of multifunctional laccase gene family in cotton (Gossypium spp.); expression and biochemical analysis during fiber development
Abstract
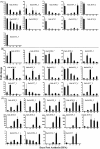
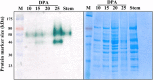
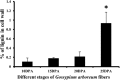
The single-celled cotton fibers, produced from seed coat epidermal cells are the largest natural source of textile fibers. The economic value of cotton fiber lies in its length and quality. The multifunctional laccase enzymes play important roles in cell elongation, lignification and pigmentation in plants and could play crucial role in cotton fiber quality. Genome-wide analysis of cultivated allotetraploid (G. hirsutum) and its progenitor diploid (G. arboreum and G. raimondii) cotton species identified 84, 44 and 46 laccase genes, respectively. Analysis of chromosomal location, phylogeny, conserved domain and physical properties showed highly conserved nature of laccases across three cotton species. Gene expression, enzymatic activity and biochemical analysis of developing cotton fibers was performed using G. arboreum species. Of the total 44, 40 laccases showed expression during different stages of fiber development. The higher enzymatic activity of laccases correlated with higher lignin content at 25 DPA (Days Post Anthesis). Further, analysis of cotton fiber phenolic compounds showed an overall decrease at 25 DPA indicating possible incorporation of these substrates into lignin polymer during secondary cell wall biosynthesis. Overall data indicate significant roles of laccases in cotton fiber development, and presents an excellent opportunity for manipulation of fiber development and quality.
Figures





Similar articles
-
Identification and analyses of miRNA genes in allotetraploid Gossypium hirsutum fiber cells based on the sequenced diploid G. raimondii genome.J Genet Genomics. 2012 Jul 20;39(7):351-60. doi: 10.1016/j.jgg.2012.04.008. Epub 2012 May 18. J Genet Genomics. 2012. PMID: 22835981
-
Genome-wide analysis of the omega-3 fatty acid desaturase gene family in Gossypium.BMC Plant Biol. 2014 Nov 18;14:312. doi: 10.1186/s12870-014-0312-5. BMC Plant Biol. 2014. PMID: 25403726 Free PMC article.
-
Metabolomic and transcriptomic insights into how cotton fiber transitions to secondary wall synthesis, represses lignification, and prolongs elongation.BMC Genomics. 2015 Jun 27;16(1):477. doi: 10.1186/s12864-015-1708-9. BMC Genomics. 2015. PMID: 26116072 Free PMC article.
-
Boosting seed development as a new strategy to increase cotton fiber yield and quality.J Integr Plant Biol. 2013 Jul;55(7):572-5. doi: 10.1111/jipb.12074. J Integr Plant Biol. 2013. PMID: 23718313 Review.
-
Transposable elements play an important role during cotton genome evolution and fiber cell development.Sci China Life Sci. 2016 Feb;59(2):112-21. doi: 10.1007/s11427-015-4928-y. Epub 2015 Dec 19. Sci China Life Sci. 2016. PMID: 26687725 Review.
Cited by
-
Identification of Grape Laccase Genes and Their Potential Role in Secondary Metabolite Synthesis.Int J Mol Sci. 2024 Sep 30;25(19):10574. doi: 10.3390/ijms251910574. Int J Mol Sci. 2024. PMID: 39408902 Free PMC article.
-
The Soybean Laccase Gene Family: Evolution and Possible Roles in Plant Defense and Stem Strength Selection.Genes (Basel). 2019 Sep 11;10(9):701. doi: 10.3390/genes10090701. Genes (Basel). 2019. PMID: 31514462 Free PMC article.
-
The role of JrLACs in the lignification of walnut endocarp.BMC Plant Biol. 2021 Nov 3;21(1):511. doi: 10.1186/s12870-021-03280-3. BMC Plant Biol. 2021. PMID: 34732134 Free PMC article.
-
The MdERF61-mdm-miR397b-MdLAC7b module regulates apple resistance to Fusarium solani via lignin biosynthesis.Plant Physiol. 2024 Dec 23;197(1):kiae518. doi: 10.1093/plphys/kiae518. Plant Physiol. 2024. PMID: 39374536 Free PMC article.
-
DkmiR397 Regulates Proanthocyanidin Biosynthesis via Negative Modulating DkLAC2 in Chinese PCNA Persimmon.Int J Mol Sci. 2022 Mar 16;23(6):3200. doi: 10.3390/ijms23063200. Int J Mol Sci. 2022. PMID: 35328620 Free PMC article.
References
-
- Gillham F. E. M. et al.. Cotton Production Prospects for the Next Decade. (The World Bank, 1996).
-
- Fan L. et al.. Molecular and Biochemical Evidence for Phenylpropanoid Synthesis and Presence of Wall-linked Phenolics in Cotton Fibers. Journal of Integrative Plant Biology 51, 626–637 (2009). - PubMed
-
- de Morais Teixeira E. et al.. Cellulose nanofibers from white and naturally colored cotton fibers. Cellulose 17, 595–606 (2010).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

