The Homeobox Transcription Factor RHOX10 Drives Mouse Spermatogonial Stem Cell Establishment
- PMID: 27681428
- PMCID: PMC5063083
- DOI: 10.1016/j.celrep.2016.08.090
The Homeobox Transcription Factor RHOX10 Drives Mouse Spermatogonial Stem Cell Establishment
Abstract
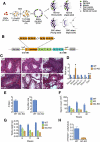
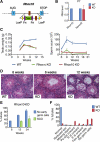
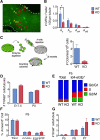
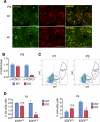
The developmental origins of most adult stem cells are poorly understood. Here, we report the identification of a transcription factor-RHOX10-critical for the initial establishment of spermatogonial stem cells (SSCs). Conditional loss of the entire 33-gene X-linked homeobox gene cluster that includes Rhox10 causes progressive spermatogenic decline, a phenotype indistinguishable from that caused by loss of only Rhox10. We demonstrate that this phenotype results from dramatically reduced SSC generation. By using a battery of approaches, including single-cell-RNA sequencing (scRNA-seq) analysis, we show that Rhox10 drives SSC generation by promoting pro-spermatogonia differentiation. Rhox10 also regulates batteries of migration genes and promotes the migration of pro-spermatogonia into the SSC niche. The identification of an X-linked homeobox gene that drives the initial generation of SSCs has implications for the evolution of X-linked gene clusters and sheds light on regulatory mechanisms influencing adult stem cell generation in general.
Keywords: Rhox; germ cell; germ line stem cell; gonocytes; homeobox; prospermatogonia; spermatogenesis; spermatogonia; spermatogonial stem cells; transcription factor.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

Comment in
-
RHOX10 Spurs on Spermatogonial Stem Cell Establishment.Biol Reprod. 2016 Dec;95(6):118. doi: 10.1095/biolreprod.116.146084. Epub 2016 Oct 19. Biol Reprod. 2016. PMID: 27908878 No abstract available.
References
-
- Almenara A, Escalante G, Gazzo E, Gonzales GF. Transillumination to evaluate spermatogenesis: effect of testosterone enanthate in adult male rats. Arch. Androl. 2001;46:21–27. - PubMed
-
- Aramaki S, Hayashi K, Kurimoto K, Ohta H, Yabuta Y, Iwanari H, Mochizuki Y, Hamakubo T, Kato Y, Shirahige K, et al. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev. Cell. 2013;27:516–529. - PubMed
-
- Basciani S, De Luca G, Dolci S, Brama M, Arizzi M, Mariani S, Rosano G, Spera G, Gnessi L. Platelet-derived growth factor receptor beta-subtype regulates proliferation and migration of gonocytes. Endocrinology. 2008;149:6226–6235. - PubMed
-
- Breen EC. VEGF in biological control. J. Cell. Biochem. 2007;102:1358–1367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

