Mutant PFN1 causes ALS phenotypes and progressive motor neuron degeneration in mice by a gain of toxicity
- PMID: 27681617
- PMCID: PMC5068285
- DOI: 10.1073/pnas.1605964113
Mutant PFN1 causes ALS phenotypes and progressive motor neuron degeneration in mice by a gain of toxicity
Abstract
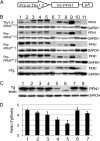
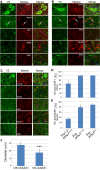
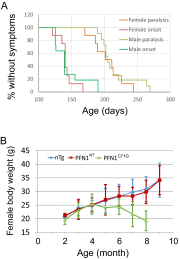
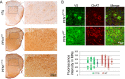
Mutations in the profilin 1 (PFN1) gene cause amyotrophic lateral sclerosis (ALS), a neurodegenerative disease caused by the loss of motor neurons leading to paralysis and eventually death. PFN1 is a small actin-binding protein that promotes formin-based actin polymerization and regulates numerous cellular functions, but how the mutations in PFN1 cause ALS is unclear. To investigate this problem, we have generated transgenic mice expressing either the ALS-associated mutant (C71G) or wild-type protein. Here, we report that mice expressing the mutant, but not the wild-type, protein had relentless progression of motor neuron loss with concomitant progressive muscle weakness ending in paralysis and death. Furthermore, mutant, but not wild-type, PFN1 forms insoluble aggregates, disrupts cytoskeletal structure, and elevates ubiquitin and p62/SQSTM levels in motor neurons. Unexpectedly, the acceleration of motor neuron degeneration precedes the accumulation of mutant PFN1 aggregates. These results suggest that although mutant PFN1 aggregation may contribute to neurodegeneration, it does not trigger its onset. Importantly, these experiments establish a progressive disease model that can contribute toward identifying the mechanisms of ALS pathogenesis and the development of therapeutic treatments.
Keywords: denervation; motor neuron disease; muscle atrophy; neurodegeneration; proteostasis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


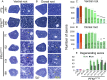
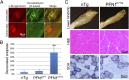
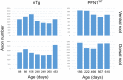
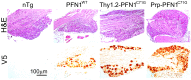

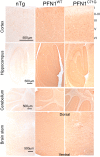
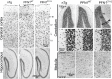


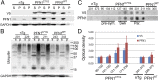
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

