Early somatic mosaicism is a rare cause of long-QT syndrome
- PMID: 27681629
- PMCID: PMC5068256
- DOI: 10.1073/pnas.1607187113
Early somatic mosaicism is a rare cause of long-QT syndrome
Abstract
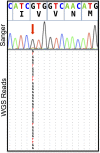
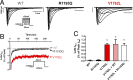
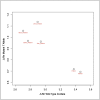
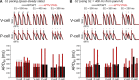
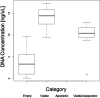
Somatic mosaicism, the occurrence and propagation of genetic variation in cell lineages after fertilization, is increasingly recognized to play a causal role in a variety of human diseases. We investigated the case of life-threatening arrhythmia in a 10-day-old infant with long QT syndrome (LQTS). Rapid genome sequencing suggested a variant in the sodium channel NaV1.5 encoded by SCN5A, NM_000335:c.5284G > T predicting p.(V1762L), but read depth was insufficient to be diagnostic. Exome sequencing of the trio confirmed read ratios inconsistent with Mendelian inheritance only in the proband. Genotyping of single circulating leukocytes demonstrated the mutation in the genomes of 8% of patient cells, and RNA sequencing of cardiac tissue from the infant confirmed the expression of the mutant allele at mosaic ratios. Heterologous expression of the mutant channel revealed significantly delayed sodium current with a dominant negative effect. To investigate the mechanism by which mosaicism might cause arrhythmia, we built a finite element simulation model incorporating Purkinje fiber activation. This model confirmed the pathogenic consequences of cardiac cellular mosaicism and, under the presenting conditions of this case, recapitulated 2:1 AV block and arrhythmia. To investigate the extent to which mosaicism might explain undiagnosed arrhythmia, we studied 7,500 affected probands undergoing commercial gene-panel testing. Four individuals with pathogenic variants arising from early somatic mutation events were found. Here we establish cardiac mosaicism as a causal mechanism for LQTS and present methods by which the general phenomenon, likely to be relevant for all genetic diseases, can be detected through single-cell analysis and next-generation sequencing.
Keywords: arrhythmia; computational modeling; genomics; mosaicism; single cell.
Conflict of interest statement
At the time of this work K.M.K., S.R., and L.B. were employed by Gilead Sciences. M.J.C., S.T.K.G., S.B., J.W., and R.C. were employed by Personalis, Inc. J.W. and E.A.A. are founders of Personalis, Inc. which offers clinical genetic testing but does not offer Clinical Laboratory Improvement Amendments (CLIA)-certified rapid-turnaround whole-genome sequencing. N.A.T. is a co-founder of CardioSolv LLC.
Figures






References
-
- Biesecker LG, Spinner NB. A genomic view of mosaicism and human disease. Nat Rev Genet. 2013;14(5):307–320. - PubMed
-
- Thibodeau IL, et al. Paradigm of genetic mosaicism and lone atrial fibrillation: Physiological characterization of a connexin 43-deletion mutant identified from atrial tissue. Circulation. 2010;122(3):236–244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

