C₁-Pathways in Methyloversatilis universalis FAM5: Genome Wide Gene Expression and Mutagenesis Studies
- PMID: 27682085
- PMCID: PMC5023235
- DOI: 10.3390/microorganisms3020175
C₁-Pathways in Methyloversatilis universalis FAM5: Genome Wide Gene Expression and Mutagenesis Studies
Abstract
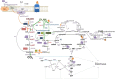
Methyloversatilis universalis FAM5 utilizes single carbon compounds such as methanol or methylamine as a sole source of carbon and energy. Expression profiling reveals distinct sets of genes altered during growth on methylamine vs methanol. As expected, all genes for the N-methylglutamate pathway were induced during growth on methylamine. Among other functions responding to the aminated source of C₁-carbon, are a heme-containing amine dehydrogenase (Qhp), a distant homologue of formaldehyde activating enzyme (Fae3), molybdenum-containing formate dehydrogenase, ferredoxin reductase, a set of homologues to urea/ammonium transporters and amino-acid permeases. Mutants lacking one of the functional subunits of the amine dehydrogenase (ΔqhpA) or Δfae3 showed no growth defect on C₁-compounds. M. universalis FAM5 strains with a lesion in the H₄-folate pathway were not able to use any C₁-compound, methanol or methylamine. Genes essential for C₁-assimilation (the serine cycle and glyoxylate shunt) and H₄MTP-pathway for formaldehyde oxidation showed similar levels of expression on both C₁-carbon sources. M. universalis FAM5 possesses three homologs of the formaldehyde activating enzyme, a key enzyme of the H₄MTP-pathway. Strains lacking the canonical Fae (fae1) lost the ability to grow on both C₁-compounds. However, upon incubation on methylamine the fae1-mutant produced revertants (Δfae1(R)), which regained the ability to grow on methylamine. Double and triple mutants (Δfae1(R)Δfae3, or Δfae1(R)Δfae2 or Δfae1(R)Δfae2Δfae3) constructed in the revertant strain background showed growth similar to the Δfae1(R) phenotype. The metabolic pathways for utilization of methanol and methylamine in Methyloversatilis universalis FAM5 are reconstructed based on these gene expression and phenotypic data.
Keywords: C1-metabolism; N-methylglutamate pathway; formaldehyde activating enzyme homologues; methyloversatilis; rhodocyclaceae.
Figures
References
-
- De Marco P., Pacheco C.C., Figueiredo A.R., Moradas-Ferreira P. Novel pollutant-resistant methylotrophic bacteria for use in bioremediation. FEMS Microbiol. Lett. 2004;234:75–80. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

