Quantitative lateral flow strip assays as User-Friendly Tools To Detect Biomarker Profiles For Leprosy
- PMID: 27682181
- PMCID: PMC5041085
- DOI: 10.1038/srep34260
Quantitative lateral flow strip assays as User-Friendly Tools To Detect Biomarker Profiles For Leprosy
Abstract
Leprosy is a debilitating, infectious disease caused by Mycobacterium leprae. Despite the availability of multidrug therapy, transmission is unremitting. Thus, early identification of M. leprae infection is essential to reduce transmission. The immune response to M. leprae is determined by host genetics, resulting in paucibacillary (PB) and multibacillary (MB) leprosy associated with dominant cellular or humoral immunity, respectively. This spectral pathology of leprosy compels detection of immunity to M. leprae to be based on multiple, diverse biomarkers. In this study we have applied quantitative user friendly lateral flow assays (LFAs) for four immune markers (anti-PGL-I antibodies, IL-10, CCL4 and IP-10) for whole blood samples from a longitudinal BCG vaccination field-trial in Bangladesh. Different biomarker profiles, in contrast to single markers, distinguished M. leprae infected from non-infected test groups, patients from household contacts (HHC) and endemic controls (EC), or MB from PB patients. The test protocol presented in this study merging detection of innate, adaptive cellular as well as humoral immunity, thus provides a convenient tool to measure specific biomarker profiles for M. leprae infection and leprosy utilizing a field-friendly technology.
Figures
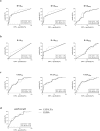

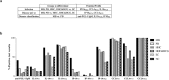

References
-
- WHO Global leprosy: update on the 2012 situation. Wkly. Epidemiol. Rec. 88, 365–379 (2013). - PubMed
-
- Lockwood D. N. & Saunderson P. Nerve damage in Leprosy: a continuing challenge for scientists, clinicians and service providers. Int Health 4, 77–85 (2012). - PubMed
-
- Geluk A. Challenges in immunodiagnostic tests for leprosy. Expert. Opin. Med. Diagn. 7, 265–274 (2013). - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

