Quantification of the Latent HIV-1 Reservoir Using Ultra Deep Sequencing and Primer ID in a Viral Outgrowth Assay
- PMID: 27683060
- PMCID: PMC5233602
- DOI: 10.1097/QAI.0000000000001187
Quantification of the Latent HIV-1 Reservoir Using Ultra Deep Sequencing and Primer ID in a Viral Outgrowth Assay
Abstract
Background: In this study, we measured the latent HIV-1 reservoir harboring replication-competent HIV-1 in resting CD4 T cells in participants on highly active antiretroviral therapy, quantitating the frequency of latent infection through the use of a Primer ID-based Ultra Deep Sequencing Assay (UDSA), in comparison to the readout of the quantitative viral outgrowth assay (QVOA).
Methods: Viral RNA derived from culture wells of QVOA that scored as HIV-1 p24 capsid antigen positive were tagged with a specific barcode during cDNA synthesis, and the sequences within the V1-V3 region of the HIV-1 env gene were analyzed for diversity using the Primer ID-based paired-end MiSeq platform. We analyzed samples from a total of 19 participants, 2 initially treated with highly active antiretroviral therapy in acute infection and 17 treated during chronic infection. Phylogenetic trees were generated with all viral lineages detected from culture wells derived from each participant to determine the number of distinct viral lineages growing out in each well, thus capturing another level of information beyond the well being positive for viral antigen. The infectious units per million (IUPM) cell values estimated using a maximum likelihood approach, based on the number of distinct viral lineages detected (VOA-UDSA), were compared with those obtained from QVOA measured using limiting dilution.
Results: IUPM estimates determined by VOA-UDSA ranged from 0.14 to 3.66 and strongly correlated with the IUPM estimates determined by QVOA (r = 0.94; P < 0.0001).
Conclusions: VOA-UDSA may be an alternative readout for that currently used for QVOA.
Conflict of interest statement
The other authors have no other conflicts of interest to disclose.
Figures


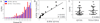

References
-
- Orenstein JM, Fox C, Wahl SM. Macrophages as a source of HIV during opportunistic infections. Science. 1997;276(5320):1857–1861. - PubMed
-
- Wightman F, Solomon A, Khoury G, et al. Both CD31(+) and CD31(−) naive CD4(+) T cells are persistent HIV type 1-infected reservoirs in individuals receiving antiretroviral therapy. J Infect Dis. 2010;202(11):1738–1748. - PubMed
-
- Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387(6629):183–188. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

