Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION
- PMID: 27685097
- PMCID: PMC5215819
- DOI: 10.1038/nprot.2016.124
Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION
Abstract
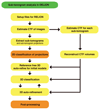
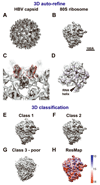
Electron cryo-tomography (cryo-ET) is a technique that is used to produce 3D pictures (tomograms) of complex objects such as asymmetric viruses, cellular organelles or whole cells from a series of tilted electron cryo-microscopy (cryo-EM) images. Averaging of macromolecular complexes found within tomograms is known as subtomogram averaging, and this technique allows structure determination of macromolecular complexes in situ. Subtomogram averaging is also gaining in popularity for the calculation of initial models for single-particle analysis. We describe herein a protocol for subtomogram averaging from cryo-ET data using the RELION software (http://www2.mrc-lmb.cam.ac.uk/relion). RELION was originally developed for cryo-EM single-particle analysis, and the subtomogram averaging approach presented in this protocol has been implemented in the existing workflow for single-particle analysis so that users may conveniently tap into existing capabilities of the RELION software. We describe how to calculate 3D models for the contrast transfer function (CTF) that describe the transfer of information in the imaging process, and we illustrate the results of classification and subtomogram averaging refinement for cryo-ET data of purified hepatitis B capsid particles and Saccharomyces cerevisiae 80S ribosomes. Using the steps described in this protocol, along with the troubleshooting and optimization guidelines, high-resolution maps can be obtained in which secondary structure elements are resolved subtomogram.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures




References
-
- Bai XC, McMullan G, Scheres SH. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40:49–57. - PubMed
-
- Baumeister W. Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr Opin Struct Biol. 2002;12:679–684. - PubMed
-
- Briggs JA. Structural biology in situ--the potential of subtomogram averaging. Curr Opin Struct Biol. 2013;23:261–267. - PubMed
-
- Beck M, Lucić V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

