Mapping 3D genome architecture through in situ DNase Hi-C
- PMID: 27685100
- PMCID: PMC5547819
- DOI: 10.1038/nprot.2016.126
Mapping 3D genome architecture through in situ DNase Hi-C
Abstract
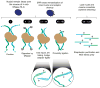
With the advent of massively parallel sequencing, considerable work has gone into adapting chromosome conformation capture (3C) techniques to study chromosomal architecture at a genome-wide scale. We recently demonstrated that the inactive murine X chromosome adopts a bipartite structure using a novel 3C protocol, termed in situ DNase Hi-C. Like traditional Hi-C protocols, in situ DNase Hi-C requires that chromatin be chemically cross-linked, digested, end-repaired, and proximity-ligated with a biotinylated bridge adaptor. The resulting ligation products are optionally sheared, affinity-purified via streptavidin bead immobilization, and subjected to traditional next-generation library preparation for Illumina paired-end sequencing. Importantly, in situ DNase Hi-C obviates the dependence on a restriction enzyme to digest chromatin, instead relying on the endonuclease DNase I. Libraries generated by in situ DNase Hi-C have a higher effective resolution than traditional Hi-C libraries, which makes them valuable in cases in which high sequencing depth is allowed for, or when hybrid capture technologies are expected to be used. The protocol described here, which involves ∼4 d of bench work, is optimized for the study of mammalian cells, but it can be broadly applicable to any cell or tissue of interest, given experimental parameter optimization.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Using DNase Hi-C techniques to map global and local three-dimensional genome architecture at high resolution.Methods. 2018 Jun 1;142:59-73. doi: 10.1016/j.ymeth.2018.01.014. Epub 2018 Jan 31. Methods. 2018. PMID: 29382556 Free PMC article.
-
Targeted DNase Hi-C.Methods Mol Biol. 2021;2157:65-83. doi: 10.1007/978-1-0716-0664-3_5. Methods Mol Biol. 2021. PMID: 32820399 Free PMC article.
-
Chromosome Conformation Capture Followed by Genome-Wide Sequencing (Hi-C) in Drosophila Embryos.Methods Mol Biol. 2023;2655:41-55. doi: 10.1007/978-1-0716-3143-0_4. Methods Mol Biol. 2023. PMID: 37212987
-
A genome-wide 3C-method for characterizing the three-dimensional architectures of genomes.Methods. 2012 Nov;58(3):277-88. doi: 10.1016/j.ymeth.2012.06.018. Epub 2012 Jul 6. Methods. 2012. PMID: 22776363 Free PMC article. Review.
-
Technical Review: A Hitchhiker's Guide to Chromosome Conformation Capture.Methods Mol Biol. 2018;1675:233-246. doi: 10.1007/978-1-4939-7318-7_14. Methods Mol Biol. 2018. PMID: 29052195 Review.
Cited by
-
Dynamic chromatin organization and regulatory interactions in human endothelial cell differentiation.Stem Cell Reports. 2023 Jan 10;18(1):159-174. doi: 10.1016/j.stemcr.2022.11.003. Epub 2022 Dec 8. Stem Cell Reports. 2023. PMID: 36493778 Free PMC article.
-
Origins and evolution of extreme life span in Pacific Ocean rockfishes.Science. 2021 Nov 12;374(6569):842-847. doi: 10.1126/science.abg5332. Epub 2021 Nov 11. Science. 2021. PMID: 34762458 Free PMC article.
-
Rapid Macrosatellite Evolution Promotes X-Linked Hybrid Male Sterility in a Feline Interspecies Cross.Mol Biol Evol. 2021 Dec 9;38(12):5588-5609. doi: 10.1093/molbev/msab274. Mol Biol Evol. 2021. PMID: 34519828 Free PMC article.
-
Novel insights into chromosomal conformations in cancer.Mol Cancer. 2017 Nov 17;16(1):173. doi: 10.1186/s12943-017-0741-5. Mol Cancer. 2017. PMID: 29149895 Free PMC article. Review.
-
Weak interactions in higher-order chromatin organization.Nucleic Acids Res. 2020 May 21;48(9):4614-4626. doi: 10.1093/nar/gkaa261. Nucleic Acids Res. 2020. PMID: 32313950 Free PMC article. Review.
References
-
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. - PubMed
-
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. - PubMed
-
- Rieder CL, Khodjakov A. Mitosis through the microscope: advances in seeing inside live dividing cells. Science. 2003;300:91–96. - PubMed
-
- BARR ML, BERTRAM EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

