The emerging role of lysine methyltransferase SETD8 in human diseases
- PMID: 27688818
- PMCID: PMC5034662
- DOI: 10.1186/s13148-016-0268-4
The emerging role of lysine methyltransferase SETD8 in human diseases
Abstract
SETD8/SET8/Pr-SET7/KMT5A is the only known lysine methyltransferase (KMT) that monomethylates lysine 20 of histone H4 (H4K20) in vivo. Lysine residues of non-histone proteins including proliferating cell nuclear antigen (PCNA) and p53 are also monomethylated. As a consequence, the methyltransferase activity of the enzyme is implicated in many essential cellular processes including DNA replication, DNA damage response, transcription modulation, and cell cycle regulation. This review aims to provide an overview of the roles of SETD8 in physiological and pathological pathways and to discuss the progress made to date in inhibiting the activity of SETD8 by small molecules, with an emphasis on their discovery, selectivity over other methyltransferases and cellular activity.
Figures
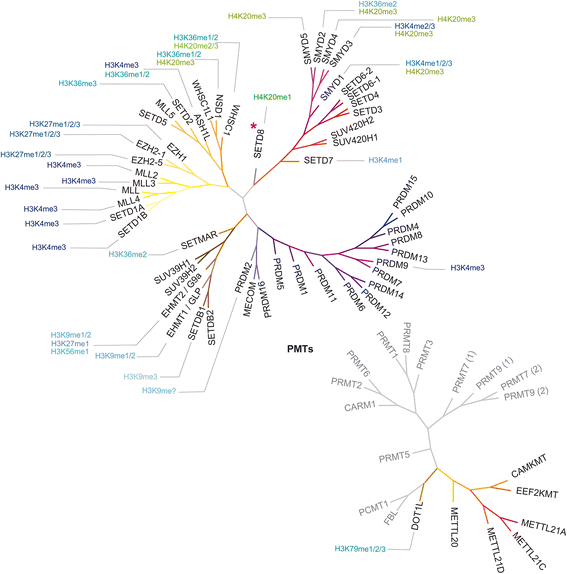
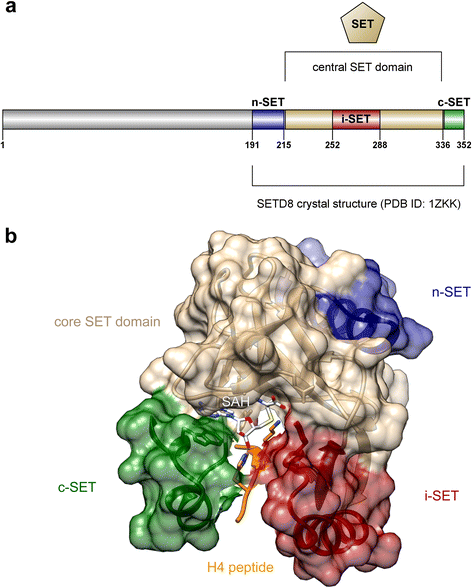
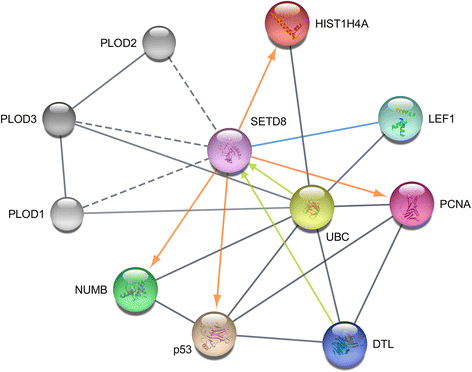
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

