DNA methylation-based measures of biological age: meta-analysis predicting time to death
- PMID: 27690265
- PMCID: PMC5076441
- DOI: 10.18632/aging.101020
DNA methylation-based measures of biological age: meta-analysis predicting time to death
Abstract
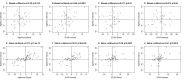
Estimates of biological age based on DNA methylation patterns, often referred to as "epigenetic age", "DNAm age", have been shown to be robust biomarkers of age in humans. We previously demonstrated that independent of chronological age, epigenetic age assessed in blood predicted all-cause mortality in four human cohorts. Here, we expanded our original observation to 13 different cohorts for a total sample size of 13,089 individuals, including three racial/ethnic groups. In addition, we examined whether incorporating information on blood cell composition into the epigenetic age metrics improves their predictive power for mortality. All considered measures of epigenetic age acceleration were predictive of mortality (p≤8.2x10-9), independent of chronological age, even after adjusting for additional risk factors (p<5.4x10-4), and within the racial/ethnic groups that we examined (non-Hispanic whites, Hispanics, African Americans). Epigenetic age estimates that incorporated information on blood cell composition led to the smallest p-values for time to death (p=7.5x10-43). Overall, this study a) strengthens the evidence that epigenetic age predicts all-cause mortality above and beyond chronological age and traditional risk factors, and b) demonstrates that epigenetic age estimates that incorporate information on blood cell counts lead to highly significant associations with all-cause mortality.
Keywords: DNA methylation; all-cause mortality; epigenetic clock; epigenetics; lifespan; mortality.
Conflict of interest statement
The Regents of the University of California is the sole owner of a provisional patent application directed at the invention of measures of epigenetic age acceleration for which SH is a named inventor. The other authors declare no conflicts of interest.
Figures



References
Publication types
MeSH terms
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- HHSN268201100008C/HL/NHLBI NIH HHS/United States
- HHSN268201100007C/HL/NHLBI NIH HHS/United States
- R01 ES025225/ES/NIEHS NIH HHS/United States
- HHSN268201100006C/HL/NHLBI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- HHSN268201100002C/WH/WHI NIH HHS/United States
- HHSN268201100007I/HL/NHLBI NIH HHS/United States
- HHSN268201100002I/HL/NHLBI NIH HHS/United States
- R01 AG042511/AG/NIA NIH HHS/United States
- HHSN268201100001C/WH/WHI NIH HHS/United States
- HHSN268201100012C/HL/NHLBI NIH HHS/United States
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- HHSN268201100001I/HL/NHLBI NIH HHS/United States
- HHSN268201100009I/HL/NHLBI NIH HHS/United States
- RF1 AG036042/AG/NIA NIH HHS/United States
- T32 GM007309/GM/NIGMS NIH HHS/United States
- HHSN268201100010C/HL/NHLBI NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- HHSN268201100005G/HL/NHLBI NIH HHS/United States
- HHSN268201100004I/HL/NHLBI NIH HHS/United States
- HHSN268201100008I/HL/NHLBI NIH HHS/United States
- HHSN268201100046C/HL/NHLBI NIH HHS/United States
- HHSN268201100011I/HL/NHLBI NIH HHS/United States
- HHSN268201100011C/HL/NHLBI NIH HHS/United States
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/M013111/1/MRC_/Medical Research Council/United Kingdom
- HHSN268201100003C/WH/WHI NIH HHS/United States
- RA/ARRA NIH HHS/United States
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- R01 ES015172/ES/NIEHS NIH HHS/United States
- R01 ES021733/ES/NIEHS NIH HHS/United States
- R01 AG042187/AG/NIA NIH HHS/United States
- P30 ES023515/ES/NIEHS NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- HHSN268201100005I/HL/NHLBI NIH HHS/United States
- HHSN271201100004C/AG/NIA NIH HHS/United States
- R01 AG029451/AG/NIA NIH HHS/United States
- HHSN268201100009C/HL/NHLBI NIH HHS/United States
- HHSN268201100005C/HL/NHLBI NIH HHS/United States
- CSO_/Chief Scientist Office/United Kingdom
- U34 AG051425/AG/NIA NIH HHS/United States
- R00 ES023450/ES/NIEHS NIH HHS/United States
- HHSN268201100004C/WH/WHI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

