Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
- PMID: 27690357
- PMCID: PMC5045296
- DOI: 10.7554/eLife.18101
Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Abstract
The kinesin-8 motor, KIF19A, accumulates at cilia tips and controls cilium length. Defective KIF19A leads to hydrocephalus and female infertility because of abnormally elongated cilia. Uniquely among kinesins, KIF19A possesses the dual functions of motility along ciliary microtubules and depolymerization of microtubules. To elucidate the molecular mechanisms of these functions we solved the crystal structure of its motor domain and determined its cryo-electron microscopy structure complexed with a microtubule. The features of KIF19A that enable its dual function are clustered on its microtubule-binding side. Unexpectedly, a destabilized switch II coordinates with a destabilized L8 to enable KIF19A to adjust to both straight and curved microtubule protofilaments. The basic clusters of L2 and L12 tether the microtubule. The long L2 with a characteristic acidic-hydrophobic-basic sequence effectively stabilizes the curved conformation of microtubule ends. Hence, KIF19A utilizes multiple strategies to accomplish the dual functions of motility and microtubule depolymerization by ATP hydrolysis.
Keywords: X-ray crystallography; biophysics; cell biology; cryo-electron microscopy; kinesin; microtubule dynamic; mouse; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures






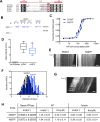

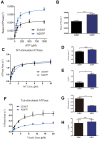
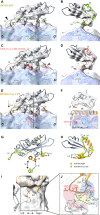


References
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. - DOI - PubMed
-
- Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SH, Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

