The 3'-untranslated region of mRNAs as a site for ribozyme cleavage-dependent processing and control in bacteria
- PMID: 27690736
- PMCID: PMC5785228
- DOI: 10.1080/15476286.2016.1240141
The 3'-untranslated region of mRNAs as a site for ribozyme cleavage-dependent processing and control in bacteria
Abstract
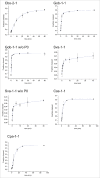
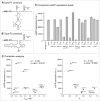
Besides its primary informational role, the sequence of the mRNA (mRNA) including its 5'- and 3'- untranslated regions (UTRs), contains important features that are relevant for post-transcriptional and translational regulation of gene expression. In this work a number of bacterial twister motifs are characterized both in vitro and in vivo. The analysis of their genetic contexts shows that these motifs have the potential of being transcribed as part of polycistronic mRNAs, thus we suggest the involvement of bacterial twister motifs in the processing of mRNA. Our data show that the ribozyme-mediated cleavage of the bacterial 3'-UTR has major effects on gene expression. While the observed effects correlate weakly with the kinetic parameters of the ribozymes, they show dependence on motif-specific structural features and on mRNA stabilization properties of the secondary structures that remain on the 3'-UTR after ribozyme cleavage. Using these principles, novel artificial twister-based riboswitches are developed that exert their activity via ligand-dependent cleavage of the 3'-UTR and the removal of the protective intrinsic terminator. Our results provide insights into possible biological functions of these recently discovered and widespread catalytic RNA motifs and offer new tools for applications in biotechnology, synthetic biology and metabolic engineering.
Keywords: Aptazyme; RNA decay; RNase; bacteria; hammerhead ribozyme; polyadenylation; riboswitch; secondary structure; twister ribozyme.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
