Convergent biosynthetic pathways to β-lactam antibiotics
- PMID: 27693891
- PMCID: PMC5161666
- DOI: 10.1016/j.cbpa.2016.09.013
Convergent biosynthetic pathways to β-lactam antibiotics
Abstract
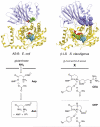
Five naturally-occurring families of β-lactams have inspired a class of drugs that constitute >60% of the antimicrobials used in human medicine. Their biosynthetic pathways reveal highly individualized synthetic strategies that yet converge on a common azetidinone ring assembled in structural contexts that confer selective binding and inhibition of d,d-transpeptidases that play essential roles in bacterial cell wall (peptidoglycan) biosynthesis. These enzymes belong to a single 'clan' of evolutionarily distinct serine hydrolases whose active site geometry and mechanism of action is specifically matched by these antibiotics for inactivation that is kinetically competitive with their native function. Unusual enzyme-mediated reactions and catalytic multitasking in these pathways are discussed with particular attention to the diverse ways the β-lactam itself is generated, and more broadly how the intrinsic reactivity of this core structural element is modulated in natural systems through the introduction of ring strain and electronic effects.
Copyright © 2016. Published by Elsevier Ltd.
Figures






References
-
- Rawlings ND, Barrett AJ, Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucl Acids Res. 2016;44:D343–D350. - PMC - PubMed
-
•• This database is extremely useful for the study of evolutionary convergence, yet tolerable differences in active site geometry for catalytic function.
-
- Buller AR, Townsend CA. Intrinsic evolutionary constraints on protease structure, enzyme acylation, and the identity of the catalytic triad. Proc Natl Acad Sci U S A. 2013;110:E653–E661. - PMC - PubMed
-
•• This article, with extensive Supplementary Information, introduces a formalism to reanalyze the reactive rotamer and oxyanion hole of all serine, cysteine and threonine proteases of independent lineage and, where possible, their inhibitors.
-
- Walsh C, Wencewitz T. Antibiotics: Challenges, Mechanisms, Opportunities. ASM Press; Washington, DC: 2016.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

