Role of lipid phosphate phosphatase 3 in human aortic endothelial cell function
- PMID: 27694435
- PMCID: PMC5157138
- DOI: 10.1093/cvr/cvw217
Role of lipid phosphate phosphatase 3 in human aortic endothelial cell function
Abstract
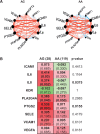
Aims: Lipid phosphate phosphatase 3; type 2 phosphatidic acid phosphatase β (LPP3; PPAP2B) is a transmembrane protein dephosphorylating and thereby terminating signalling of lipid substrates including lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P). Human LPP3 possesses a cell adhesion motif that allows interaction with integrins. A polymorphism (rs17114036) in PPAP2B is associated with coronary artery disease, which prompted us to investigate the possible role of LPP3 in human endothelial dysfunction, a condition promoting atherosclerosis.
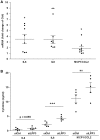
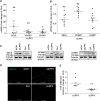
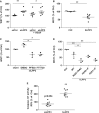
Methods and results: To study the role of LPP3 in endothelial cells we used human primary aortic endothelial cells (HAECs) in which LPP3 was silenced or overexpressed using either wild type or mutated cDNA constructs. LPP3 silencing in HAECs enhanced secretion of inflammatory cytokines, leucocyte adhesion, cell survival, and migration and impaired angiogenesis, whereas wild-type LPP3 overexpression reversed these effects and induced apoptosis. We also demonstrated that LPP3 expression was negatively correlated with vascular endothelial growth factor expression. Mutations in either the catalytic or the arginine-glycine-aspartate (RGD) domains impaired endothelial cell function and pharmacological inhibition of S1P or LPA restored it. LPA was not secreted in HAECs under silencing or overexpressing LPP3. However, the intra- and extra-cellular levels of S1P tended to be correlated with LPP3 expression, indicating that S1P is probably degraded by LPP3.
Conclusions: We demonstrated that LPP3 is a negative regulator of inflammatory cytokines, leucocyte adhesion, cell survival, and migration in HAECs, suggesting a protective role of LPP3 against endothelial dysfunction in humans. Both the catalytic and the RGD functional domains were involved and S1P, but not LPA, might be the endogenous substrate of LPP3.
Keywords: Angiogenesis; Apoptosis; Atherosclerosis; Endothelial dysfunction.
Published on behalf of the European Society of Cardiology. All rights reserved. © The Author 2016. For Permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Hopkins PN. Molecular biology of atherosclerosis. Physiol Rev 2013;93:1317–1542. - PubMed
-
- Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AFR, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Cardiogenics, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Mühleisen TW, Muhlestein JB, Münzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nöthen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schäfer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, März W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O'Donnell CJ, McPherson R, Erdmann J, CARDIoGRAM Consortium, Samani NJ. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–338. - PMC - PubMed
-
- Reschen ME, Gaulton KJ, Lin D, Soilleux EJ, Morris AJ, Smyth SS, O’Callaghan CA. Lipid-induced epigenomic changes in human macrophages identify a coronary artery disease-associated variant that regulates PPAP2B expression through altered C/EBP-beta binding. PLoS Genet 2015;11:e1005061. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

