Histone variant H3F3A promotes lung cancer cell migration through intronic regulation
- PMID: 27694942
- PMCID: PMC5477500
- DOI: 10.1038/ncomms12914
Histone variant H3F3A promotes lung cancer cell migration through intronic regulation
Abstract
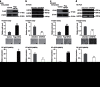
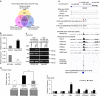
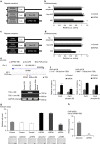
Although several somatic single nucleotide variations in histone H3.3 have been investigated as cancer drivers, other types of aberration have not been well studied. Here, we demonstrate that overexpression of H3F3A, encoding H3.3, is associated with lung cancer progression and promotes lung cancer cell migration by activating metastasis-related genes. H3.3 globally activates gene expression through the occupation of intronic regions in lung cancer cells. Moreover, H3.3 binding regions show characteristics of regulatory DNA elements. We show that H3.3 is deposited at a specific intronic region of GPR87, where it modifies the chromatin status and directly activates GPR87 transcription. The expression levels of H3F3A and GPR87, either alone or in combination, are robust prognostic markers for early-stage lung cancer, and may indicate potential for the development of treatments involving GPR87 antagonists. In summary, our results demonstrate that intronic regulation by H3F3A may be a target for the development of novel therapeutic strategies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Siegel R., Naishadham D. & Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30 (2013). - PubMed
-
- Hoffman P. C., Mauer A. M. & Vokes E. E. Lung cancer. Lancet 355, 479–485 (2000). - PubMed
-
- Nesbitt J. C., Putnam J. B. Jr., Walsh G. L., Roth J. A. & Mountain C. F. Survival in early-stage non-small cell lung cancer. Ann. Thorac. Surg. 60, 466–472 (1995). - PubMed
-
- Roselli M. et al.. Postsurgical chemotherapy in stage IB nonsmall cell lung cancer: long-term survival in a randomized study. Int. J. Cancer 119, 955–960 (2006). - PubMed
-
- Douillard J. Y. et al.. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol. 7, 719–727 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

