Mutations in the HECT domain of NEDD4L lead to AKT-mTOR pathway deregulation and cause periventricular nodular heterotopia
- PMID: 27694961
- PMCID: PMC5086093
- DOI: 10.1038/ng.3676
Mutations in the HECT domain of NEDD4L lead to AKT-mTOR pathway deregulation and cause periventricular nodular heterotopia
Abstract
Neurodevelopmental disorders with periventricular nodular heterotopia (PNH) are etiologically heterogeneous, and their genetic causes remain in many cases unknown. Here we show that missense mutations in NEDD4L mapping to the HECT domain of the encoded E3 ubiquitin ligase lead to PNH associated with toe syndactyly, cleft palate and neurodevelopmental delay. Cellular and expression data showed sensitivity of PNH-associated mutants to proteasome degradation. Moreover, an in utero electroporation approach showed that PNH-related mutants and excess wild-type NEDD4L affect neurogenesis, neuronal positioning and terminal translocation. Further investigations, including rapamycin-based experiments, found differential deregulation of pathways involved. Excess wild-type NEDD4L leads to disruption of Dab1 and mTORC1 pathways, while PNH-related mutations are associated with deregulation of mTORC1 and AKT activities. Altogether, these data provide insights into the critical role of NEDD4L in the regulation of mTOR pathways and their contributions in cortical development.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
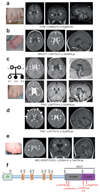
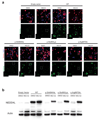

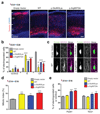
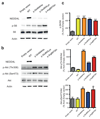
References
-
- Caviness VS, Jr, Takahashi T, Nowakowski RS. Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci. 1995;18:379–83. - PubMed
-
- Rakic P, Caviness VS., Jr Cortical development: view from neurological mutants two decades later. Neuron. 1995;14:1101–4. - PubMed
-
- Francis F, et al. Human disorders of cortical development: from past to present. Eur J Neurosci. 2006;23:877–93. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

