A novel glycan modifies the flagellar filament proteins of the oral bacterium Treponema denticola
- PMID: 27696564
- PMCID: PMC5182079
- DOI: 10.1111/mmi.13544
A novel glycan modifies the flagellar filament proteins of the oral bacterium Treponema denticola
Abstract
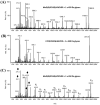
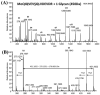
While protein glycosylation has been reported in several spirochetes including the syphilis bacterium Treponema pallidum and Lyme disease pathogen Borrelia burgdorferi, the pertinent glycan structures and their roles remain uncharacterized. Herein, a novel glycan with an unusual chemical composition and structure in the oral spirochete Treponema denticola, a keystone pathogen of periodontitis was reported. The identified glycan of mass 450.2 Da is composed of a monoacetylated nonulosonic acid (Non) with a novel extended N7 acyl modification, a 2-methoxy-4,5,6-trihydroxy-hexanoyl residue in which the Non has a pseudaminic acid configuration (L-glycero-L-manno) and is β-linked to serine or threonine residues. This novel glycan modifies the flagellin proteins (FlaBs) of T. denticola by O-linkage at multiple sites near the D1 domain, a highly conserved region of bacterial flagellins that interact with Toll-like receptor 5. Furthermore, mutagenesis studies demonstrate that the glycosylation plays an essential role in the flagellar assembly and motility of T. denticola. To our knowledge, this novel glycan and its unique modification sites have not been reported previously in any bacteria.
© 2016 John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures


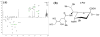





Similar articles
-
Identification and Characterization of the Alternative σ28 Factor in Treponema denticola.J Bacteriol. 2022 Sep 20;204(9):e0024822. doi: 10.1128/jb.00248-22. Epub 2022 Aug 31. J Bacteriol. 2022. PMID: 36043861 Free PMC article.
-
Transcriptional and functional characterizations of multiple flagellin genes in spirochetes.Mol Microbiol. 2022 Sep;118(3):175-190. doi: 10.1111/mmi.14959. Epub 2022 Jul 18. Mol Microbiol. 2022. PMID: 35776658 Free PMC article.
-
Anti-σ28 Factor FlgM Regulates Flagellin Gene Expression and Flagellar Polarity of Treponema denticola.J Bacteriol. 2023 Feb 22;205(2):e0046322. doi: 10.1128/jb.00463-22. Epub 2023 Jan 30. J Bacteriol. 2023. PMID: 36715541 Free PMC article.
-
Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications.J Mol Microbiol Biotechnol. 2006;11(3-5):167-91. doi: 10.1159/000094053. J Mol Microbiol Biotechnol. 2006. PMID: 16983194 Review.
-
Gram-negative flagella glycosylation.Int J Mol Sci. 2014 Feb 19;15(2):2840-57. doi: 10.3390/ijms15022840. Int J Mol Sci. 2014. PMID: 24557579 Free PMC article. Review.
Cited by
-
Structural and Biosynthetic Diversity of Nonulosonic Acids (NulOs) That Decorate Surface Structures in Bacteria.Trends Microbiol. 2021 Feb;29(2):142-157. doi: 10.1016/j.tim.2020.08.002. Epub 2020 Sep 17. Trends Microbiol. 2021. PMID: 32950378 Free PMC article. Review.
-
Flagellin Glycoproteomics of the Periodontitis Associated Pathogen Selenomonas sputigena Reveals Previously Not Described O-glycans and Rhamnose Fragment Rearrangement Occurring on the Glycopeptides.Mol Cell Proteomics. 2018 Apr;17(4):721-736. doi: 10.1074/mcp.RA117.000394. Epub 2018 Jan 16. Mol Cell Proteomics. 2018. PMID: 29339411 Free PMC article.
-
Identification and Characterization of the Alternative σ28 Factor in Treponema denticola.J Bacteriol. 2022 Sep 20;204(9):e0024822. doi: 10.1128/jb.00248-22. Epub 2022 Aug 31. J Bacteriol. 2022. PMID: 36043861 Free PMC article.
-
Analysis of a flagellar filament cap mutant reveals that HtrA serine protease degrades unfolded flagellin protein in the periplasm of Borrelia burgdorferi.Mol Microbiol. 2019 Jun;111(6):1652-1670. doi: 10.1111/mmi.14243. Epub 2019 Apr 26. Mol Microbiol. 2019. PMID: 30883947 Free PMC article.
-
Transcriptional and functional characterizations of multiple flagellin genes in spirochetes.Mol Microbiol. 2022 Sep;118(3):175-190. doi: 10.1111/mmi.14959. Epub 2022 Jul 18. Mol Microbiol. 2022. PMID: 35776658 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

