Proprotein Convertase Processing Enhances Peroxidasin Activity to Reinforce Collagen IV
- PMID: 27697841
- PMCID: PMC5104926
- DOI: 10.1074/jbc.M116.745935
Proprotein Convertase Processing Enhances Peroxidasin Activity to Reinforce Collagen IV
Abstract
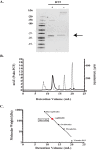
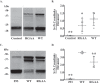
The basement membrane (BM) is a form of extracellular matrix that underlies cell layers in nearly all animal tissues. Type IV collagen, a major constituent of BMs, is critical for tissue development and architecture. The enzyme peroxidasin (Pxdn), an extracellular matrix-associated protein, catalyzes the formation of structurally reinforcing sulfilimine cross-links within the collagen IV network, an event essential to basement membrane integrity. Although the catalytic function of Pxdn is known, the regulation of its activity remains unclear. In this work we show through N-terminal sequencing, pharmacologic studies, and mutational analysis that proprotein convertases (PCs) proteolytically process human Pxdn at Arg-1336, a location relatively close to its C terminus. PC processing enhances the enzymatic activity of Pxdn and facilitates the formation of sulfilimine cross-links in collagen IV. Thus, PC processing of Pxdn is a key regulatory step that contributes to its function and, therefore, supports BM integrity and homeostasis.
Keywords: basement membrane; collagen; collagen IV; extracellular matrix; furin; peroxidase; peroxidasin; proprotein convertase; sulfilimine bond.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
The role of peroxidasin in solid cancer progression.Biochem Soc Trans. 2023 Oct 31;51(5):1881-1895. doi: 10.1042/BST20230018. Biochem Soc Trans. 2023. PMID: 37801286 Free PMC article. Review.
-
The Ancient Immunoglobulin Domains of Peroxidasin Are Required to Form Sulfilimine Cross-links in Collagen IV.J Biol Chem. 2015 Aug 28;290(35):21741-8. doi: 10.1074/jbc.M115.673996. Epub 2015 Jul 15. J Biol Chem. 2015. PMID: 26178375 Free PMC article.
-
Characterization of the Proprotein Convertase-Mediated Processing of Peroxidasin and Peroxidasin-like Protein.Antioxidants (Basel). 2021 Sep 30;10(10):1565. doi: 10.3390/antiox10101565. Antioxidants (Basel). 2021. PMID: 34679700 Free PMC article.
-
The sulfilimine cross-link of collagen IV contributes to kidney tubular basement membrane stiffness.Am J Physiol Renal Physiol. 2017 Sep 1;313(3):F596-F602. doi: 10.1152/ajprenal.00096.2017. Epub 2017 Apr 19. Am J Physiol Renal Physiol. 2017. PMID: 28424209 Free PMC article.
-
Role of Hypohalous Acids in Basement Membrane Homeostasis.Antioxid Redox Signal. 2017 Oct 20;27(12):839-854. doi: 10.1089/ars.2017.7245. Epub 2017 Jul 31. Antioxid Redox Signal. 2017. PMID: 28657332 Free PMC article. Review.
Cited by
-
Peroxidasin-mediated bromine enrichment of basement membranes.Proc Natl Acad Sci U S A. 2020 Jul 7;117(27):15827-15836. doi: 10.1073/pnas.2007749117. Epub 2020 Jun 22. Proc Natl Acad Sci U S A. 2020. PMID: 32571911 Free PMC article.
-
Proteolytic processing of lysyl oxidase-like-2 in the extracellular matrix is required for crosslinking of basement membrane collagen IV.J Biol Chem. 2017 Oct 13;292(41):16970-16982. doi: 10.1074/jbc.M117.798603. Epub 2017 Sep 1. J Biol Chem. 2017. PMID: 28864775 Free PMC article.
-
Pre-steady-state Kinetics Reveal the Substrate Specificity and Mechanism of Halide Oxidation of Truncated Human Peroxidasin 1.J Biol Chem. 2017 Mar 17;292(11):4583-4592. doi: 10.1074/jbc.M117.775213. Epub 2017 Jan 31. J Biol Chem. 2017. PMID: 28154175 Free PMC article.
-
The role of peroxidasin in solid cancer progression.Biochem Soc Trans. 2023 Oct 31;51(5):1881-1895. doi: 10.1042/BST20230018. Biochem Soc Trans. 2023. PMID: 37801286 Free PMC article. Review.
-
Measuring peroxidasin activity in live cells using bromide addition for signal amplification.Redox Biol. 2022 Aug;54:102385. doi: 10.1016/j.redox.2022.102385. Epub 2022 Jun 30. Redox Biol. 2022. PMID: 35803124 Free PMC article.
References
-
- Pöschl E., Schlötzer-Schrehardt U., Brachvogel B., Saito K., Ninomiya Y., and Mayer U. (2004) Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development 131, 1619–1628 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

