Nuclei migrate through constricted spaces using microtubule motors and actin networks in C. elegans hypodermal cells
- PMID: 27697906
- PMCID: PMC5117218
- DOI: 10.1242/dev.141192
Nuclei migrate through constricted spaces using microtubule motors and actin networks in C. elegans hypodermal cells
Abstract
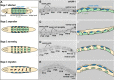
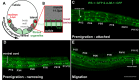
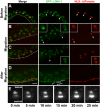
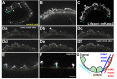
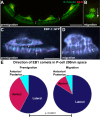
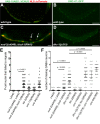
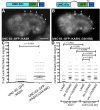
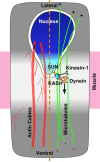
Cellular migrations through constricted spaces are a crucial aspect of many developmental and disease processes including hematopoiesis, inflammation and metastasis. A limiting factor in these events is nuclear deformation. Here, we establish an in vivo model in which nuclei can be visualized while moving through constrictions and use it to elucidate mechanisms for nuclear migration. C. elegans hypodermal P-cell larval nuclei traverse a narrow space that is about 5% their width. This constriction is blocked by fibrous organelles, structures that pass through P cells to connect the muscles to cuticle. Fibrous organelles are removed just prior to nuclear migration, when nuclei and lamins undergo extreme morphological changes to squeeze through the space. Both actin and microtubule networks are organized to mediate nuclear migration. The LINC complex, consisting of the SUN protein UNC-84 and the KASH protein UNC-83, recruits dynein and kinesin-1 to the nuclear surface. Both motors function in P-cell nuclear migration, but dynein, functioning through UNC-83, plays a more central role as nuclei migrate towards minus ends of polarized microtubule networks. Thus, the nucleoskeleton and cytoskeleton are coordinated to move nuclei through constricted spaces.
Keywords: C. elegans; Dynein; KASH; Nuclear migration; SUN.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures
References
-
- Altun Z. F. and Hall D. H. (2009a). Epithelial system, hypodermis. WormAtlas, doi:10.3908/wormatlas.1.13 10.3908/wormatlas.1.13 - DOI
-
- Altun Z. F. and Hall D. H. (2009b). Muscle system, somatic muscle. WormAtlas, doi:10.3908/wormatlas.1.7 10.3908/wormatlas.1.7 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

