ASB7 regulates spindle dynamics and genome integrity by targeting DDA3 for proteasomal degradation
- PMID: 27697924
- PMCID: PMC5057283
- DOI: 10.1083/jcb.201603062
ASB7 regulates spindle dynamics and genome integrity by targeting DDA3 for proteasomal degradation
Abstract
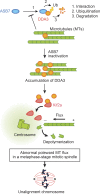
Proper dynamic regulation of the spindle is essential for successful cell division. However, the molecular mechanisms that regulate spindle dynamics in mitosis are not fully understood. In this study, we show that Cullin 5-interacting suppressor of cytokine signaling box protein ASB7 ubiquitinates DDA3, a regulator of spindle dynamics, thereby targeting it for proteasomal degradation. The presence of microtubules (MTs) prevented the ASB7-DDA3 interaction, thus stabilizing DDA3. Knockdown of ASB7 decreased MT polymerization and increased the proportion of cells with unaligned chromosomes, and this phenotype was rescued by deletion of DDA3. Collectively, these data indicate that ASB7 plays a crucial role in regulating spindle dynamics and genome integrity by controlling the expression of DDA3.
© 2016 Uematsu et al.
Figures





Similar articles
-
DDA3 and Mdp3 modulate Kif2a recruitment onto the mitotic spindle to control minus-end spindle dynamics.J Cell Sci. 2016 Jul 15;129(14):2719-25. doi: 10.1242/jcs.180109. Epub 2016 Jun 9. J Cell Sci. 2016. PMID: 27284004
-
ANKRD53 interacts with DDA3 and regulates chromosome integrity during mitosis.Biochem Biophys Res Commun. 2016 Feb 12;470(3):484-491. doi: 10.1016/j.bbrc.2016.01.144. Epub 2016 Jan 25. Biochem Biophys Res Commun. 2016. PMID: 26820536
-
DDA3 targets Cep290 into the centrosome to regulate spindle positioning.Biochem Biophys Res Commun. 2015 Jul 17-24;463(1-2):88-94. doi: 10.1016/j.bbrc.2015.05.028. Epub 2015 May 18. Biochem Biophys Res Commun. 2015. PMID: 25998387
-
Cell cycle regulation of Rho signaling pathways.Cell Cycle. 2012 Aug 15;11(16):3003-10. doi: 10.4161/cc.21088. Epub 2012 Jul 24. Cell Cycle. 2012. PMID: 22825247 Free PMC article. Review.
-
The role of 26S proteasome-dependent proteolysis in the formation and restructuring of microtubule networks.Plant Signal Behav. 2012 Oct 1;7(10):1289-95. doi: 10.4161/psb.21543. Epub 2012 Aug 20. Plant Signal Behav. 2012. PMID: 22902696 Free PMC article. Review.
Cited by
-
ASB17 Facilitates the Burst of LPS-Induced Inflammation Through Maintaining TRAF6 Stability.Front Cell Infect Microbiol. 2022 Jan 31;12:759077. doi: 10.3389/fcimb.2022.759077. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35174103 Free PMC article.
-
ASB7 Is a Novel Regulator of Cytoskeletal Organization During Oocyte Maturation.Front Cell Dev Biol. 2020 Nov 5;8:595917. doi: 10.3389/fcell.2020.595917. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33251222 Free PMC article.
-
The functions and properties of cullin-5, a potential therapeutic target for cancers.Am J Transl Res. 2020 Feb 15;12(2):618-632. eCollection 2020. Am J Transl Res. 2020. PMID: 32194910 Free PMC article. Review.
-
Molecular insights into degron recognition by CRL5ASB7 ubiquitin ligase.Nat Commun. 2024 Jul 22;15(1):6177. doi: 10.1038/s41467-024-50556-x. Nat Commun. 2024. PMID: 39039081 Free PMC article.
-
High Expression of PSRC1 Predicts Poor Prognosis in Lung Adenocarcinoma.J Cancer. 2023 Oct 7;14(17):3321-3334. doi: 10.7150/jca.88635. eCollection 2023. J Cancer. 2023. PMID: 37928428 Free PMC article.
References
-
- Eagleson G., Pfister K., Knowlton A.L., Skoglund P., Keller R., and Stukenberg P.T.. 2015. Kif2a depletion generates chromosome segregation and pole coalescence defects in animal caps and inhibits gastrulation of the Xenopus embryo. Mol. Biol. Cell. 26:924–937. 10.1091/mbc.E13-12-0721 - DOI - PMC - PubMed
-
- Gaedcke J., Grade M., Jung K., Camps J., Jo P., Emons G., Gehoff A., Sax U., Schirmer M., Becker H., et al. . 2010. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer. 49:1024–1034. 10.1002/gcc.20811 - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

