Identification of Functions Affecting Predator-Prey Interactions between Myxococcus xanthus and Bacillus subtilis
- PMID: 27698086
- PMCID: PMC5116937
- DOI: 10.1128/JB.00575-16
Identification of Functions Affecting Predator-Prey Interactions between Myxococcus xanthus and Bacillus subtilis
Abstract
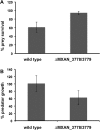
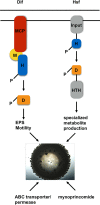
Soil bacteria engage each other in competitive and cooperative ways to determine their microenvironments. In this study, we report the identification of a large number of genes required for Myxococcus xanthus to engage Bacillus subtilis in a predator-prey relationship. We generated and tested over 6,000 individual transposon insertion mutants of M. xanthus and found many new factors required to promote efficient predation, including the specialized metabolite myxoprincomide, an ATP-binding cassette (ABC) transporter permease, and a clustered regularly interspaced short palindromic repeat (CRISPR) locus encoding bacterial immunity. We also identified genes known to be involved in predation, including those required for the production of exopolysaccharides and type IV pilus (T4P)-dependent motility, as well as chemosensory and two-component systems. Furthermore, deletion of these genes confirmed their role during predation. Overall, M. xanthus predation appears to be a multifactorial process, with multiple determinants enhancing predation capacity.
Importance: Soil bacteria engage each other in complex environments and utilize multiple traits to ensure survival. Here, we report the identification of multiple traits that enable a common soil organism, Myxococcus xanthus, to prey upon and utilize nutrients from another common soil organism, Bacillus subtilis We mutagenized the predator and carried out a screen to identify genes that were required to either enhance or diminish capacity to consume prey. We identified dozens of genes encoding factors that contribute to the overall repertoire for the predator to successfully engage its prey in the natural environment.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

