Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses
- PMID: 27698118
- PMCID: PMC5098629
- DOI: 10.1073/pnas.1607152113
Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses
Abstract
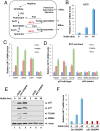
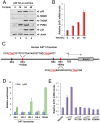
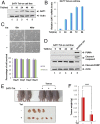
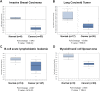
Although p53-mediated cell-cycle arrest, senescence, and apoptosis remain critical barriers to cancer development, the emerging role of p53 in cell metabolism, oxidative responses, and ferroptotic cell death has been a topic of great interest. Nevertheless, it is unclear how p53 orchestrates its activities in multiple metabolic pathways into tumor suppressive effects. Here, we identified the SAT1 (spermidine/spermine N1-acetyltransferase 1) gene as a transcription target of p53. SAT1 is a rate-limiting enzyme in polyamine catabolism critically involved in the conversion of spermidine and spermine back to putrescine. Surprisingly, we found that activation of SAT1 expression induces lipid peroxidation and sensitizes cells to undergo ferroptosis upon reactive oxygen species (ROS)-induced stress, which also leads to suppression of tumor growth in xenograft tumor models. Notably, SAT1 expression is down-regulated in human tumors, and CRISPR-cas9-mediated knockout of SAT1 expression partially abrogates p53-mediated ferroptosis. Moreover, SAT1 induction is correlated with the expression levels of arachidonate 15-lipoxygenase (ALOX15), and SAT1-induced ferroptosis is significantly abrogated in the presence of PD146176, a specific inhibitor of ALOX15. Thus, our findings uncover a metabolic target of p53 involved in ferroptotic cell death and provide insight into the regulation of polyamine metabolism and ferroptosis-mediated tumor suppression.
Keywords: SAT1; ferroptosis; p53; polyamine metabolism; tumor suppression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
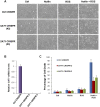
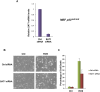
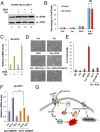

Comment in
-
Ironing out how p53 regulates ferroptosis.Proc Natl Acad Sci U S A. 2016 Nov 1;113(44):12350-12352. doi: 10.1073/pnas.1615159113. Epub 2016 Oct 17. Proc Natl Acad Sci U S A. 2016. PMID: 27791175 Free PMC article. No abstract available.
Similar articles
-
Cerebroprotein hydrolysate-I ameliorates cognitive dysfunction in APP/PS1 mice by inhibiting ferroptosis via the p53/SAT1/ALOX15 signalling pathway.Eur J Pharmacol. 2024 Sep 15;979:176820. doi: 10.1016/j.ejphar.2024.176820. Epub 2024 Jul 18. Eur J Pharmacol. 2024. PMID: 39032765
-
Depletion of the polyamines spermidine and spermine by overexpression of spermidine/spermine N¹-acetyltransferase 1 (SAT1) leads to mitochondria-mediated apoptosis in mammalian cells.Biochem J. 2015 Jun 15;468(3):435-47. doi: 10.1042/BJ20150168. Epub 2015 Apr 7. Biochem J. 2015. PMID: 25849284 Free PMC article.
-
Lipoxygenase inhibitors protect acute lymphoblastic leukemia cells from ferroptotic cell death.Biochem Pharmacol. 2017 Sep 15;140:41-52. doi: 10.1016/j.bcp.2017.06.112. Epub 2017 Jun 6. Biochem Pharmacol. 2017. PMID: 28595877
-
Spermidine/spermine-N(1)-acetyltransferase: a key metabolic regulator.Am J Physiol Endocrinol Metab. 2008 Jun;294(6):E995-1010. doi: 10.1152/ajpendo.90217.2008. Epub 2008 Mar 18. Am J Physiol Endocrinol Metab. 2008. PMID: 18349109 Review.
-
The tumor suppressor protein p53 and the ferroptosis network.Free Radic Biol Med. 2019 Mar;133:162-168. doi: 10.1016/j.freeradbiomed.2018.05.074. Epub 2018 May 23. Free Radic Biol Med. 2019. PMID: 29800655 Free PMC article. Review.
Cited by
-
Role of ferroptosis pathways in neuroinflammation and neurological disorders: From pathogenesis to treatment.Heliyon. 2024 Jan 19;10(3):e24786. doi: 10.1016/j.heliyon.2024.e24786. eCollection 2024 Feb 15. Heliyon. 2024. PMID: 38314277 Free PMC article. Review.
-
JNK inhibitor and ferroptosis modulator as possible therapeutic modalities in Alzheimer disease (AD).Sci Rep. 2024 Oct 7;14(1):23293. doi: 10.1038/s41598-024-73596-1. Sci Rep. 2024. PMID: 39375359 Free PMC article.
-
Ferroptosis: Emerging mechanisms, biological function, and therapeutic potential in cancer and inflammation.Cell Death Discov. 2024 Jan 24;10(1):45. doi: 10.1038/s41420-024-01825-7. Cell Death Discov. 2024. PMID: 38267442 Free PMC article. Review.
-
Breast cancer cells have an increased ferroptosis risk induced by system xc- blockade after deliberately downregulating CYTL1 to mediate malignancy.Redox Biol. 2024 Apr;70:103034. doi: 10.1016/j.redox.2024.103034. Epub 2024 Jan 6. Redox Biol. 2024. PMID: 38211443 Free PMC article.
-
Ferroptosis in radiation-induced brain injury: roles and clinical implications.Biomed Eng Online. 2024 Sep 11;23(1):93. doi: 10.1186/s12938-024-01288-y. Biomed Eng Online. 2024. PMID: 39261942 Free PMC article. Review.
References
-
- Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137(3):413–431. - PubMed
-
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. - PubMed
-
- Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–16. - PubMed
-
- Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

