Emodin Exerts an Antiapoptotic Effect on Human Chronic Myelocytic Leukemia K562 Cell Lines by Targeting the PTEN/PI3K-AKT Signaling Pathway and Deleting BCR-ABL
- PMID: 27698265
- PMCID: PMC5739139
- DOI: 10.1177/1534735416664784
Emodin Exerts an Antiapoptotic Effect on Human Chronic Myelocytic Leukemia K562 Cell Lines by Targeting the PTEN/PI3K-AKT Signaling Pathway and Deleting BCR-ABL
Abstract
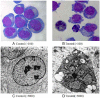
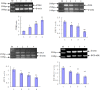
The BCR-ABL kinase inhibitor, imatinib mesylate, is the front-line treatment for chronic myeloid leukemia, but the emergence of imatinib resistance has led to the search for alternative drug treatments. There is a pressing need, therefore, to develop and test novel drugs. Natural products including plants, microorganisms, and halobios provide rich resources for discovery of anticancer drugs. In this article, we demonstrate that emodin inhibited the growth of K562 cells harboring BCR-ABL in vitro and in vivo, and induced abundant apoptosis, which was correlated with the inhibition of PETN/PI3K/Akt level and deletion of BCR-ABL. These findings suggest that emodin is a promising agent to kill K562 cells harboring BCR-ABL.
Keywords: BCR-ABL; K562 cells; PI3K-AKT; PTEN; emodin.
Conflict of interest statement
Figures









Similar articles
-
EPS8 regulates proliferation, apoptosis and chemosensitivity in BCR-ABL positive cells via the BCR-ABL/PI3K/AKT/mTOR pathway.Oncol Rep. 2018 Jan;39(1):119-128. doi: 10.3892/or.2017.6102. Epub 2017 Nov 20. Oncol Rep. 2018. PMID: 29192326 Free PMC article.
-
Emodin Inhibits Resistance to Imatinib by Downregulation of Bcr-Abl and STAT5 and Allosteric Inhibition in Chronic Myeloid Leukemia Cells.Biol Pharm Bull. 2020;43(10):1526-1533. doi: 10.1248/bpb.b20-00325. Biol Pharm Bull. 2020. PMID: 32999163
-
Inducible SHP-2 activation confers resistance to imatinib in drug-tolerant chronic myeloid leukemia cells.Toxicol Appl Pharmacol. 2018 Dec 1;360:249-256. doi: 10.1016/j.taap.2018.09.044. Epub 2018 Oct 2. Toxicol Appl Pharmacol. 2018. PMID: 30290167
-
[MET/ERK and MET/JNK Pathway Activation Is Involved in BCR-ABL Inhibitor-resistance in Chronic Myeloid Leukemia].Yakugaku Zasshi. 2018;138(12):1461-1466. doi: 10.1248/yakushi.18-00142. Yakugaku Zasshi. 2018. PMID: 30504658 Review. Japanese.
-
Combating TKI resistance in CML by inhibiting the PI3K/Akt/mTOR pathway in combination with TKIs: a review.Med Oncol. 2021 Jan 16;38(1):10. doi: 10.1007/s12032-021-01462-5. Med Oncol. 2021. PMID: 33452624 Review.
Cited by
-
[CRISPR/Cas9-mediated microRNA-21 knockout increased imatinib sensitivity in chronic myeloid leukemia cells].Zhonghua Xue Ye Xue Za Zhi. 2021 Mar 14;42(3):243-249. doi: 10.3760/cma.j.issn.0253-2727.2021.03.011. Zhonghua Xue Ye Xue Za Zhi. 2021. PMID: 33910311 Free PMC article. Chinese.
-
The Impact of Oxidative Stress and AKT Pathway on Cancer Cell Functions and Its Application to Natural Products.Antioxidants (Basel). 2022 Sep 19;11(9):1845. doi: 10.3390/antiox11091845. Antioxidants (Basel). 2022. PMID: 36139919 Free PMC article. Review.
-
Emodin-induced autophagy against cell apoptosis through the PI3K/AKT/mTOR pathway in human hepatocytes.Drug Des Devel Ther. 2019 Sep 3;13:3171-3180. doi: 10.2147/DDDT.S204958. eCollection 2019. Drug Des Devel Ther. 2019. PMID: 31564833 Free PMC article.
-
Emodin promotes fibroblast apoptosis and prevents epidural fibrosis through PERK pathway in rats.J Orthop Surg Res. 2019 Oct 10;14(1):319. doi: 10.1186/s13018-019-1357-9. J Orthop Surg Res. 2019. PMID: 31601256 Free PMC article.
-
Emodin reverses 5-Fu resistance in human colorectal cancer via downregulation of PI3K/Akt signaling pathway.Am J Transl Res. 2020 May 15;12(5):1851-1861. eCollection 2020. Am J Transl Res. 2020. PMID: 32509181 Free PMC article.
References
-
- Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, McCubrey JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18:189-218. - PubMed
-
- Olsen BB, Bjørling-Poulsen M, Guerra B. Emodin negatively affects the phosphoinositide 3-kinase/AKT signalling pathway: a study on its mechanism of action. Int J Biochem Cell Biol. 2007;39:227-237. - PubMed
-
- Fresno Vara JA, Casado E, Castro J, Cejas P, Belda-Iniesta C, González-Barón M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30:193-204. - PubMed
-
- Razis E, Bobos M, Kotoula V, et al. Evaluation of the association of PIK3CA mutations and PTEN loss with efficacy of trastuzumab therapy in metastatic breast cancer. Breast Cancer Res Treat. 2011;128:447-456. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

