Survival Motor Neuron (SMN) protein is required for normal mouse liver development
- PMID: 27698380
- PMCID: PMC5048144
- DOI: 10.1038/srep34635
Survival Motor Neuron (SMN) protein is required for normal mouse liver development
Erratum in
-
Corrigendum: Survival Motor Neuron (SMN) protein is required for normal mouse liver development.Sci Rep. 2016 Nov 10;6:35898. doi: 10.1038/srep35898. Sci Rep. 2016. PMID: 27830828 Free PMC article. No abstract available.
Abstract
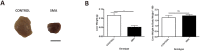
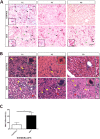
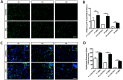
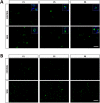
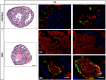
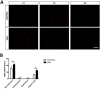
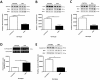
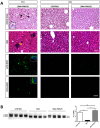
Spinal Muscular Atrophy (SMA) is caused by mutation or deletion of the survival motor neuron 1 (SMN1) gene. Decreased levels of, cell-ubiquitous, SMN protein is associated with a range of systemic pathologies reported in severe patients. Despite high levels of SMN protein in normal liver, there is no comprehensive study of liver pathology in SMA. We describe failed liver development in response to reduced SMN levels, in a mouse model of severe SMA. The SMA liver is dark red, small and has: iron deposition; immature sinusoids congested with blood; persistent erythropoietic elements and increased immature red blood cells; increased and persistent megakaryocytes which release high levels of platelets found as clot-like accumulations in the heart. Myelopoiesis in contrast, was unaffected. Further analysis revealed significant molecular changes in SMA liver, consistent with the morphological findings. Antisense treatment from birth with PMO25, increased lifespan and ameliorated all morphological defects in liver by postnatal day 21. Defects in the liver are evident at birth, prior to motor system pathology, and impair essential liver function in SMA. Liver is a key recipient of SMA therapies, and systemically delivered antisense treatment, completely rescued liver pathology. Liver therefore, represents an important therapeutic target in SMA.
Conflict of interest statement
FM is a principal investigator on Ionis funded clinical trial on AON in SMA; and in two Roche funded trials on SMA. Since 2014 he is member of the Pfizer Rare Disease Scientific Advisory Board. The remaining authors declare no competing financial interests.
Figures
References
-
- Werdnig G. Two early infantile hereditary cases of progressive muscular atrophy simulating dystrophy, but on a neural basis. Arch. Psychiat. Nurs. 22, 437–481 (1891). - PubMed
-
- Hoffmann J. Familial spinal muscular atrophy in infancy. Dtsch. Z. Nervenheilkd. 3, 427–470 (1892).
-
- Lefebvre S. et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 80, 155–165 (1995). - PubMed
-
- Hamilton G. & Gillingwater T. H. Spinal muscular atrophy: going beyond the motor neuron. Trends. Mol. Med. 19, 40–50 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

