Deubiquitinase activity is required for the proteasomal degradation of misfolded cytosolic proteins upon heat-stress
- PMID: 27698423
- PMCID: PMC5059457
- DOI: 10.1038/ncomms12907
Deubiquitinase activity is required for the proteasomal degradation of misfolded cytosolic proteins upon heat-stress
Abstract
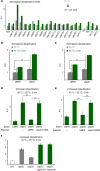
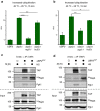
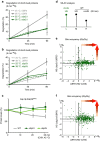
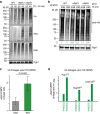
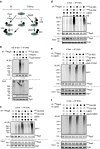
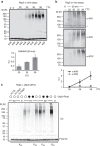
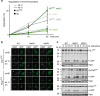
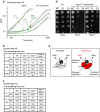
Elimination of misfolded proteins is crucial for proteostasis and to prevent proteinopathies. Nedd4/Rsp5 emerged as a major E3-ligase involved in multiple quality control pathways that target misfolded plasma membrane proteins, aggregated polypeptides and cytosolic heat-induced misfolded proteins for degradation. It remained unclear how in one case cytosolic heat-induced Rsp5 substrates are destined for proteasomal degradation, whereas other Rsp5 quality control substrates are otherwise directed to lysosomal degradation. Here we find that Ubp2 and Ubp3 deubiquitinases are required for the proteasomal degradation of cytosolic misfolded proteins targeted by Rsp5 after heat-shock (HS). The two deubiquitinases associate more with Rsp5 upon heat-stress to prevent the assembly of K63-linked ubiquitin on Rsp5 heat-induced substrates. This activity was required to promote the K48-mediated proteasomal degradation of Rsp5 HS-induced substrates. Our results indicate that ubiquitin chain editing is key to the cytosolic protein quality control under stress conditions.
Figures
Similar articles
-
Rsp5/Nedd4 is the main ubiquitin ligase that targets cytosolic misfolded proteins following heat stress.Nat Cell Biol. 2014 Dec;16(12):1227-37. doi: 10.1038/ncb3054. Epub 2014 Oct 26. Nat Cell Biol. 2014. PMID: 25344756 Free PMC article.
-
Deubiquitinating enzymes Ubp2 and Ubp15 regulate endocytosis by limiting ubiquitination and degradation of ARTs.Mol Biol Cell. 2017 May 1;28(9):1271-1283. doi: 10.1091/mbc.E17-01-0008. Epub 2017 Mar 15. Mol Biol Cell. 2017. PMID: 28298493 Free PMC article.
-
Adaptor linked K63 di-ubiquitin activates Nedd4/Rsp5 E3 ligase.Elife. 2022 Jun 30;11:e77424. doi: 10.7554/eLife.77424. Elife. 2022. PMID: 35770973 Free PMC article.
-
Hul5 ubiquitin ligase: good riddance to bad proteins.Prion. 2012 Jul 1;6(3):240-4. doi: 10.4161/pri.19929. Epub 2012 Jul 1. Prion. 2012. PMID: 22561164 Free PMC article. Review.
-
Ubiquitin and endocytic protein sorting.Essays Biochem. 2005;41:81-98. doi: 10.1042/EB0410081. Essays Biochem. 2005. PMID: 16250899 Review.
Cited by
-
Deubiquitylating enzymes in neuronal health and disease.Cell Death Dis. 2021 Jan 22;12(1):120. doi: 10.1038/s41419-020-03361-5. Cell Death Dis. 2021. PMID: 33483467 Free PMC article. Review.
-
Variable repeats in the eukaryotic polyubiquitin gene ubi4 modulate proteostasis and stress survival.Nat Commun. 2017 Aug 30;8(1):397. doi: 10.1038/s41467-017-00533-4. Nat Commun. 2017. PMID: 28855501 Free PMC article.
-
It's not just a phase; ubiquitination in cytosolic protein quality control.Biochem Soc Trans. 2021 Feb 26;49(1):365-377. doi: 10.1042/BST20200694. Biochem Soc Trans. 2021. PMID: 33634825 Free PMC article. Review.
-
K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains.Proc Natl Acad Sci U S A. 2018 Feb 13;115(7):E1401-E1408. doi: 10.1073/pnas.1716673115. Epub 2018 Jan 29. Proc Natl Acad Sci U S A. 2018. PMID: 29378950 Free PMC article.
-
Hsp40/70/110 chaperones adapt nuclear protein quality control to serve cytosolic clients.J Cell Biol. 2018 Jun 4;217(6):2019-2032. doi: 10.1083/jcb.201706091. Epub 2018 Apr 13. J Cell Biol. 2018. PMID: 29653997 Free PMC article.
References
-
- Comyn S. A., Chan G. T. & Mayor T. False start: cotranslational protein ubiquitination and cytosolic protein quality control. J. Proteomics 100, 92–101 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

