Comparative and parallel genome-wide association studies for metabolic and agronomic traits in cereals
- PMID: 27698483
- PMCID: PMC5059443
- DOI: 10.1038/ncomms12767
Comparative and parallel genome-wide association studies for metabolic and agronomic traits in cereals
Abstract
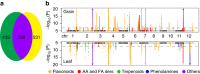
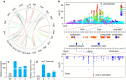
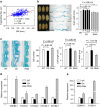
The plant metabolome is characterized by extensive diversity and is often regarded as a bridge between genome and phenome. Here we report metabolic and phenotypic genome-wide studies (mGWAS and pGWAS) in rice grain that, in addition to previous metabolic GWAS in rice leaf and maize kernel, show both distinct and overlapping aspects of genetic control of metabolism within and between species. We identify new candidate genes potentially influencing important metabolic and/or morphological traits. We show that the differential genetic architecture of rice metabolism between different tissues is in part determined by tissue specific expression. Using parallel mGWAS and pGWAS we identify new candidate genes potentially responsible for variation in traits such as grain colour and size, and provide evidence of metabotype-phenotype linkage. Our study demonstrates a powerful strategy for interactive functional genomics and metabolomics in plants, especially the cloning of minor QTLs for complex phenotypic traits.
Figures

References
-
- Keurentjes J. J. Genetical metabolomics: closing in on phenotypes. Curr. Opin. Plant Biol. 12, 223–230 (2009). - PubMed
-
- Saito K. & Matsuda F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 61, 463–489 (2010). - PubMed
-
- Fiehn O. et al.. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18, 1157–1161 (2000). - PubMed
-
- Keurentjes J. J. et al.. The genetics of plant metabolism. Nat. Genet. 38, 842–849 (2006). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

