Insights from lncRNAs Profiling of MIN6 Beta Cells Undergoing Inflammation
- PMID: 27698546
- PMCID: PMC5028877
- DOI: 10.1155/2016/9275106
Insights from lncRNAs Profiling of MIN6 Beta Cells Undergoing Inflammation
Abstract
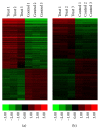
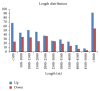
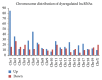
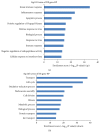
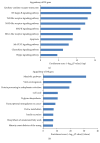
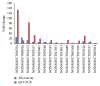
Type 1 diabetes mellitus (T1DM) is an organ-specific autoimmune disease characterized by chronic and progressive apoptotic destruction of pancreatic beta cells. During the initial phases of T1DM, cytokines and other inflammatory mediators released by immune cells progressively infiltrate islet cells, induce alterations in gene expression, provoke functional impairment, and ultimately lead to apoptosis. Long noncoding RNAs (lncRNAs) are a new important class of pervasive genes that have a variety of biological functions and play key roles in many diseases. However, whether they have a function in cytokine-induced beta cell apoptosis is still uncertain. In this study, lncRNA microarray technology was used to identify the differently expressed lncRNAs and mRNAs in MIN6 cells exposed to proinflammatory cytokines. Four hundred forty-four upregulated and 279 downregulated lncRNAs were detected with a set filter fold-change ≧2.0. To elucidate the potential functions of these lncRNAs, Gene Ontology (GO) and pathway analyses were used to evaluate the potential functions of differentially expressed lncRNAs. Additionally, a lncRNA-mRNA coexpression network was constructed to predict the interactions between the most strikingly regulated lncRNAs and mRNAs. This study may be utilized as a background or reference resource for future functional studies on lncRNAs related to the diagnosis and development of new therapies for T1DM.
Figures
References
-
- Patterson C. C., Dahlquist G. G., Gyürüs E., Green A., Soltész G. Incidence trends for childhood type 1 diabetes in Europe during 1989–2003 and predicted new cases 2005–2020: a multicentre prospective registration study. The Lancet. 2009;373(9680):2027–2033. doi: 10.1016/s0140-6736(09)60568-7. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

