Significant loss of sensitivity and specificity in the taxonomic classification occurs when short 16S rRNA gene sequences are used
- PMID: 27699286
- PMCID: PMC5037269
- DOI: 10.1016/j.heliyon.2016.e00170
Significant loss of sensitivity and specificity in the taxonomic classification occurs when short 16S rRNA gene sequences are used
Abstract
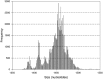
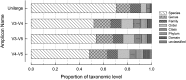
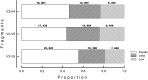
The classification performance of Kraken was evaluated in terms of sensitivity and specificity when using short and long 16S rRNA sequences. A total of 440,738 sequences from bacteria with complete taxonomic classifications were downloaded from the high quality ribosomal RNA database SILVA. Amplicons produced (86,371 sequences; 1450 bp) by virtual PCR with primers covering the V1-V9 region of the 16S-rRNA gene were used as reference. Virtual PCŔs of internal fragments V3-V4, V4-V5 and V3-V5 were performed. A total of 81,523, 82,334 and 82,998 amplicons were obtained for regions V3-V4, V4-V5 and V3-V5 respectively. Differences in depth of taxonomic classification were detected among the internal fragments. For instance, sensitivity and specificity of sequences classified up to subspecies level were higher when the largest internal fraction (V3-V5) was used (54.0 and 74.6% respectively), compared to V3-V4 (45.1 and 66.7%) and V4-V5 (41.8 and 64.6%) fragments. Similar pattern was detected for sequences classified up to more superficial taxonomic categories (i.e. family, order, class…). Results also demonstrate that internal fragments lost specificity and some could be misclassified at the deepest taxonomic levels (i.e. species or subspecies). It is concluded that the larger V3-V5 fragment could be considered for massive high throughput sequencing reducing the loss of sensitivity and sensibility.
Keywords: Bioinformatics; Biological sciences; Genetics; Microbiology.
Figures
References
-
- Baker G.C., Smith J.J., Cowan D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods. 2003;55:541–555. - PubMed
-
- Baldi P., Brunak S., Chauvin Y., Andersen C.A.F., Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

