Paucibacterial Microbiome and Resident DNA Virome of the Healthy Conjunctiva
- PMID: 27699405
- PMCID: PMC5054734
- DOI: 10.1167/iovs.16-19803
Paucibacterial Microbiome and Resident DNA Virome of the Healthy Conjunctiva
Abstract
Purpose: To characterize the ocular surface microbiome of healthy volunteers using a combination of microbial culture and high-throughput DNA sequencing techniques.
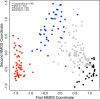
Methods: Conjunctival swab samples from 107 healthy volunteers were analyzed by bacterial culture, 16S rDNA gene deep sequencing (n = 89), and biome representational in silico karyotyping (BRiSK; n = 80). Swab samples of the facial skin (n = 42), buccal mucosa (n = 50), and environmental controls (n = 27) were processed in parallel. 16S rDNA gene quantitative PCR was used to calculate the bacterial load in each site. Bacteria were characterized by site using principal coordinate analysis of metagenomics data. BRiSK data were analyzed for presence of fungi and viruses.
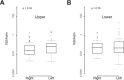
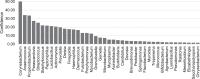
Results: Corynebacteria, Propionibacteria, and coagulase-negative Staphylococci were the predominant organisms identified by all three techniques. Quantitative 16S PCR demonstrated approximately 0.1 bacterial 16S rDNA/human actin copy on the ocular surface compared with greater than 10 16S rDNA/human actin copy for facial skin or the buccal mucosa. The conjunctival bacterial community structure is distinct compared with the facial skin (R = 0.474, analysis of similarities P = 0.0001), the buccal mucosa (R = 0.893, P = 0.0001), and environmental control samples (R = 0.536, P = 0.0001). 16S metagenomics revealed substantially more bacterial diversity on the ocular surface than other techniques, which appears to be artifactual. BRiSK revealed presence of torque teno virus (TTV) on the healthy ocular surface, which was confirmed by direct PCR to be present in 65% of all conjunctiva samples tested.
Conclusions: Relative to adjacent skin or other mucosa, healthy ocular surface microbiome is paucibacterial. Its flora are distinct from adjacent skin. Torque teno virus is a frequent constituent of the ocular surface microbiome. (ClinicalTrials.gov number, NCT02298881.).
Figures


References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

