Laboratory evolution of artificially expanded DNA gives redesignable aptamers that target the toxic form of anthrax protective antigen
- PMID: 27701076
- PMCID: PMC5175368
- DOI: 10.1093/nar/gkw890
Laboratory evolution of artificially expanded DNA gives redesignable aptamers that target the toxic form of anthrax protective antigen
Abstract
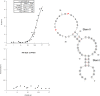
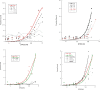
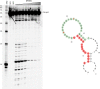
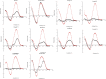
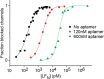
Reported here is a laboratory in vitro evolution (LIVE) experiment based on an artificially expanded genetic information system (AEGIS). This experiment delivers the first example of an AEGIS aptamer that binds to an isolated protein target, the first whose structural contact with its target has been outlined and the first to inhibit biologically important activities of its target, the protective antigen from Bacillus anthracis We show how rational design based on secondary structure predictions can also direct the use of AEGIS to improve the stability and binding of the aptamer to its target. The final aptamer has a dissociation constant of ∼35 nM. These results illustrate the value of AEGIS-LIVE for those seeking to obtain receptors and ligands without the complexities of medicinal chemistry, and also challenge the biophysical community to develop new tools to analyze the spectroscopic signatures of new DNA folds that will emerge in synthetic genetic systems replacing standard DNA and RNA as platforms for LIVE.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. - PubMed
-
- Ellington A.D., Szostak J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. - PubMed
-
- Battersby T.R., Ang D.N., Burgstaller P., Jurczyk S.C., Bowser M.T., Buchanan D.D., Kennedy R.T., Benner S.A. Quantitative analysis of receptors for adenosine nucleotides obtained via in vitro selection from a library incorporating a cationic nucleotide analog. J. Am. Chem. Soc. 1999;121:9781–9789. - PubMed
-
- Jäger S., Rasched G., Kornreich-Leshem H., Engeser M., Thum O., Famulok M. A versatile toolbox for variable DNA functionalization at high density. J. Am. Chem. Soc. 2005;127:15071–15082. - PubMed
-
- Hollenstein M., Hipolito C.J., Lam C.H., Perrin D.M. A DNAzyme with three protein-like functional groups: enhancing catalytic efficiency of M2+-independent RNA cleavage. Chembiochem. 2009;10:1988–1992. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

