LINEs between Species: Evolutionary Dynamics of LINE-1 Retrotransposons across the Eukaryotic Tree of Life
- PMID: 27702814
- PMCID: PMC5203782
- DOI: 10.1093/gbe/evw243
LINEs between Species: Evolutionary Dynamics of LINE-1 Retrotransposons across the Eukaryotic Tree of Life
Abstract
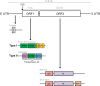
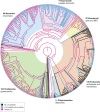
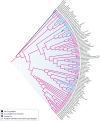
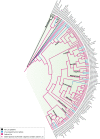
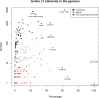
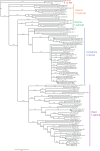
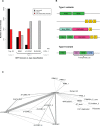
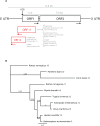
LINE-1 (L1) retrotransposons are dynamic elements. They have the potential to cause great genomic change because of their ability to 'jump' around the genome and amplify themselves, resulting in the duplication and rearrangement of regulatory DNA. Active L1, in particular, are often thought of as tightly constrained, homologous and ubiquitous elements with well-characterized domain organization. For the past 30 years, model organisms have been used to define L1s as 6-8 kb sequences containing a 5'-UTR, two open reading frames working harmoniously in cis, and a 3'-UTR with a polyA tail. In this study, we demonstrate the remarkable and overlooked diversity of L1s via a comprehensive phylogenetic analysis of elements from over 500 species from widely divergent branches of the tree of life. The rapid and recent growth of L1 elements in mammalian species is juxtaposed against the diverse lineages found in other metazoans and plants. In fact, some of these previously unexplored mammalian species (e.g. snub-nosed monkey, minke whale) exhibit L1 retrotranspositional 'hyperactivity' far surpassing that of human or mouse. In contrast, non-mammalian L1s have become so varied that the current classification system seems to inadequately capture their structural characteristics. Our findings illustrate how both long-term inherited evolutionary patterns and random bursts of activity in individual species can significantly alter genomes, highlighting the importance of L1 dynamics in eukaryotes.
Keywords: LINE; eukaryotes; evolution; retrotransposon; transposable element.
© The Author(s) 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Adelson DL, Raison JM, Garber M, Edgar RC. 2010. Interspersed repeats in the horse (Equus caballus); spatial correlations highlight conserved chromosomal domains. Anim Genet. 41 (Suppl 2):91–99. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

